6JOX
 
 | | triosephosphate isomerase-scylla paramamosain | | Descriptor: | Triosephosphate isomerase | | Authors: | Xia, F, Jin, T. | | Deposit date: | 2019-03-25 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal Structure Analysis and Conformational Epitope Mutation of Triosephosphate Isomerase, a Mud Crab Allergen.
J.Agric.Food Chem., 67, 2019
|
|
7N8U
 
 | |
7ABX
 
 | | Perdeuterated E65Q-TIM complexed with 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kelpsas, V, Caldararu, O, von Wachenfeldt, C, Oksanen, E. | | Deposit date: | 2020-09-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Neutron structures of Leishmania mexicana triosephosphate isomerase in complex with reaction-intermediate mimics shed light on the proton-shuttling steps.
Iucrj, 8, 2021
|
|
1HG3
 
 | | Crystal structure of tetrameric TIM from Pyrococcus woesei. | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Walden, H, Bell, G.S, Russell, R.J.M, Siebers, B, Hensel, R, Taylor, G.L. | | Deposit date: | 2000-12-12 | | Release date: | 2001-03-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tiny Tim: A Small, Tetrameric, Hyperthermostable Triosephosphate Isomerase
J.Mol.Biol., 306, 2001
|
|
8UZ7
 
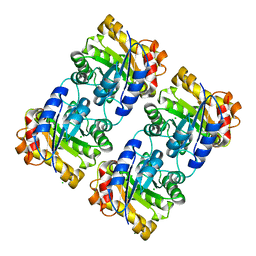 | | Crystal structure of a novel triose phosphate isomerase identified on the shrimp transcriptome | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Triosephosphate isomerase | | Authors: | Sotelo-Mundo, R.R, Gomez-Yanes, A.C, Lopez-Zavala, A.A, Lopez-Garcia, J.A, Ochoa-Leyva, A. | | Deposit date: | 2023-11-14 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a novel triose phosphate isomerase from shrimp
To Be Published
|
|
8TIM
 
 | |
1CI1
 
 | | CRYSTAL STRUCTURE OF TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI IN HEXANE | | Descriptor: | HEXANE, PROTEIN (TRIOSEPHOSPHATE ISOMERASE) | | Authors: | Gao, X.-G, Maldondo, E, Perez-Montfort, R, De Gomez-Puyou, M.T, Gomez-Puyou, A, Rodriguez-Romero, A. | | Deposit date: | 1999-04-06 | | Release date: | 1999-09-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of triosephosphate isomerase from Trypanosoma cruzi in hexane.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4WJE
 
 | |
1DKW
 
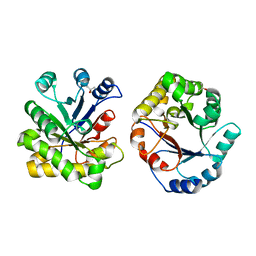 | | CRYSTAL STRUCTURE OF TRIOSE-PHOSPHATE ISOMERASE WITH MODIFIED SUBSTRATE BINDING SITE | | Descriptor: | TERTIARY-BUTYL ALCOHOL, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Norledge, B.V, Lambeir, A.M, Abagyan, R.A, Rottman, A, Fernandez, A.M, Filimonov, V.V, Peter, M.G, Wierenga, R.K. | | Deposit date: | 1999-12-08 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Modeling, mutagenesis, and structural studies on the fully conserved phosphate-binding loop (loop 8) of triosephosphate isomerase: toward a new substrate specificity.
Proteins, 42, 2001
|
|
6R8H
 
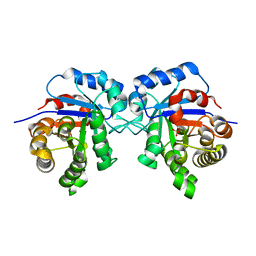 | | Triosephosphate isomerase from liver fluke (Fasciola hepatica). | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Ferraro, F, Corvo, I, Bergalli, L, Ilarraz, A, Cabrera, M, Gil, J, Susuki, B, Caffrey, C, Timson, D.J, Robert, X, Guillon, C, Alvarez, G. | | Deposit date: | 2019-04-01 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel and selective inactivators of Triosephosphate isomerase with anti-trematode activity.
Sci Rep, 10, 2020
|
|
4X22
 
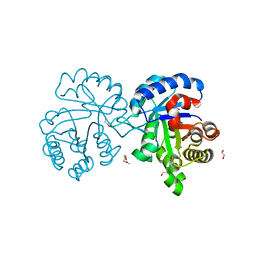 | | Crystal structure of Leptospira Interrogans Triosephosphate Isomerase (LiTIM) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
2BTM
 
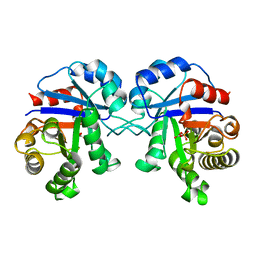 | |
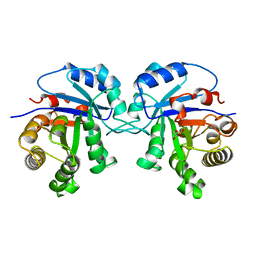
6TIM
 
 | |
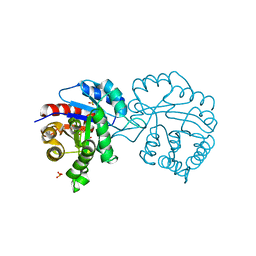
2DP3
 
 | |
7UXV
 
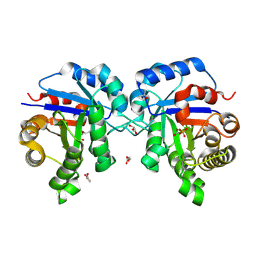 | | human triosephosphate isomerase mutant G122R | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Romero, J.M. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | human triosephosphate isomerase mutant G122R
To Be Published
|
|
7UXB
 
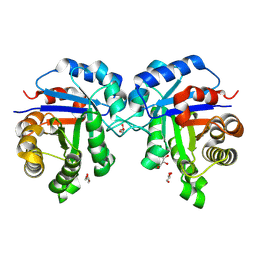 | | Human triosephosphate isomerase mutant G122R | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Triosephosphate isomerase | | Authors: | Romero, J.M. | | Deposit date: | 2022-05-05 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human triosephosphate isomerase mutant G122R
To Be Published
|
|
7V03
 
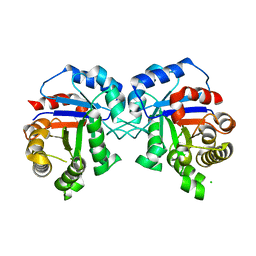 | |
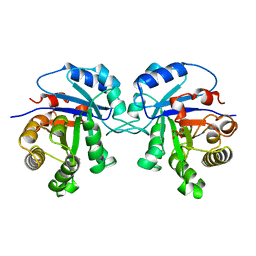
4TIM
 
 | |
4UNK
 
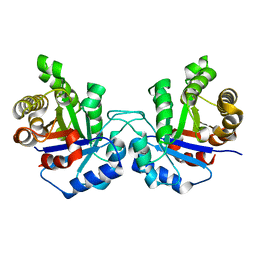 | | Crystal structure of human triosephosphate isomerase (mutant N15D) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | DeLaMora-DeLaMora, I, Torres-Larios, A, Enriquez-Flores, S, Mendez, S.T, Castillo-Villanueva, A, Gomez-Manzo, S, Lopez-Velazquez, G, Marcial-Quino, J, Torres-Arroyo, A, Garcia-Torres, I, Reyes-Vivas, H, Oria-Hernandez, J. | | Deposit date: | 2014-05-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Triosephosphate Isomerase (Mutant N15D)
To be Published
|
|
4UNL
 
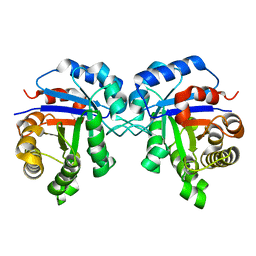 | | Crystal structure of a single mutant (N71D) of triosephosphate isomerase from human | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | DeLaMora-DeLaMora, I, Torres-Larios, A, Enriquez-Flores, S, Mendez, S.T, Castillo-Villanueva, A, Gomez-Manzo, S, Lopez-Velazquez, G, Marcial-Quino, J, Torres-Arroyo, A, Garcia-Torres, I, Reyes-Vivas, H, Oria-Hernandez, J. | | Deposit date: | 2014-05-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Single Mutant (N71D) of Triosephosphate Isomerase from Human
To be Published
|
|
5EYW
 
 | | Crystal structure of Litopenaeus vannamei triosephosphate isomerase complexed with 2-Phosphoglycolic acid | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Carrasco-Miranda, J.S, Lopez-Zavala, A.A, Brieba, L.G, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2015-11-25 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights from a novel invertebrate triosephosphate isomerase from Litopenaeus vannamei.
Biochim.Biophys.Acta, 1864, 2016
|
|
5GV4
 
 | |
5GZP
 
 | |
3GVG
 
 | |
5TIM
 
 | | REFINED 1.83 ANGSTROMS STRUCTURE OF TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE, CRYSTALLIZED IN THE PRESENCE OF 2.4 M-AMMONIUM SULPHATE. A COMPARISON WITH THE STRUCTURE OF THE TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE-GLYCEROL-3-PHOSPHATE COMPLEX | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Wierenga, R.K, Hol, W.G.J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex.
J.Mol.Biol., 220, 1991
|
|
