6H8S
 
 | |
7S4F
 
 | | Protein Tyrosine Phosphatase 1B - F182Q mutant bound with Hepes | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brandao, T.A.S, Hengge, A.C, Johnson, S.J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
7XC0
 
 | | Crystal structure of Human RPTPH | | Descriptor: | PHOSPHATE ION, Receptor-type tyrosine-protein phosphatase H | | Authors: | Kim, M, Ryu, S.E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the catalytic domain of human RPTPH.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6SUC
 
 | | Human PTPRU D1 domain, oxidised form | | Descriptor: | Receptor-type tyrosine-protein phosphatase U | | Authors: | Hay, I.M, Fearnley, G.W, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2019-09-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The receptor PTPRU is a redox sensitive pseudophosphatase.
Nat Commun, 11, 2020
|
|
6SUB
 
 | | Human PTPRU D1 domain, reduced form | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase U | | Authors: | Hay, I.M, Fearnley, G.W, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2019-09-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The receptor PTPRU is a redox sensitive pseudophosphatase.
Nat Commun, 11, 2020
|
|
7TVJ
 
 | | Crystal Structure of Monobody Mb(SHP2PTP_13)/SHP2 PTP Domain Complex | | Descriptor: | CITRATE ANION, Mb(SHP2PTP_13), Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sha, F, Koide, S. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Monobody Inhibitor Selective to the Phosphatase Domain of SHP2 and its Use as a Probe for Quantifying SHP2 Allosteric Regulation.
J.Mol.Biol., 435, 2023
|
|
7UAD
 
 | | Crystal structure of human PTPN2 with inhibitor ABBV-CLS-484 | | Descriptor: | 5-{(7R)-1-fluoro-3-hydroxy-7-[(3-methylbutyl)amino]-5,6,7,8-tetrahydronaphthalen-2-yl}-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Longenecker, K.L, Qiu, W, Sun, Q, Frost, J.M. | | Deposit date: | 2022-03-12 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | The PTPN2/PTPN1 inhibitor ABBV-CLS-484 unleashes potent anti-tumour immunity.
Nature, 622, 2023
|
|
6UZT
 
 | | Crystal Structure of RPTP alpha | | Descriptor: | Receptor-type tyrosine-protein phosphatase alpha | | Authors: | Santelli, E, Wen, Y, Yang, S, Svensson, M.N.D, Stanford, S.M, Bottini, N. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPTP alpha phosphatase activity is allosterically regulated by the membrane-distal catalytic domain.
J.Biol.Chem., 295, 2020
|
|
6W30
 
 | | Protein Tyrosine Phosphatase 1B Bound to Amorphadiene | | Descriptor: | Amorphadiene, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sarkar, A, Kim, E.Y, Hongdusit, A, Sankaran, B, Fox, J. | | Deposit date: | 2020-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Microbially Guided Discovery and Biosynthesis of Biologically Active Natural Products.
Acs Synth Biol, 10, 2021
|
|
8CT8
 
 | | Crystal structure of Drosophila melanogaster PRL/CBS-pair domain complex | | Descriptor: | IODIDE ION, PRL-1 phosphatase, Unextended protein | | Authors: | Fakih, R, Goldstein, R.H, Kozlov, G, Gehring, K. | | Deposit date: | 2022-05-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Burst kinetics and CNNM binding are evolutionarily conserved properties of phosphatases of regenerating liver.
J.Biol.Chem., 299, 2023
|
|
8DU7
 
 | | Room-temperature serial synchrotron crystallography (SSX) structure of apo PTP1B | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Sharma, S, Keedy, D.A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room-temperature serial synchrotron crystallography of the human phosphatase PTP1B.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
5H08
 
 | | Human PTPRZ D1 domain complexed with NAZ2329 | | Descriptor: | 3-{[2-Ethoxy-5-(trifluoromethyl)benzyl]sulfanyl}-N-(phenylsulfonyl)thiophene-2-carboxamide, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Sugawara, H. | | Deposit date: | 2016-10-04 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Targeting PTPRZ inhibits stem cell-like properties and tumorigenicity in glioblastoma cells
Sci Rep, 7, 2017
|
|
5HDE
 
 | | Crystal Structure of PTPN12 Catalytic Domain | | Descriptor: | PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Dong, H, Li, S, Shi, J. | | Deposit date: | 2016-01-05 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of PTPN12 Catalytic Domain
To Be Published
|
|
4BJO
 
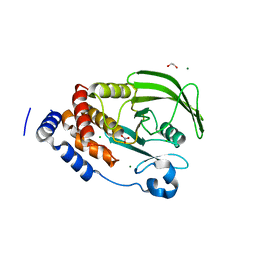 | | Nitrate in the active site of PTP1b is a putative mimetic of the transition state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kenny, P.W, Newman, J, Peat, T.S. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Nitrate in the Active Site of Protein Tyrosine Phosphatase 1B is a Putative Mimetic of the Transition State.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZ1
 
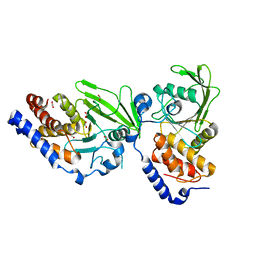 | | Crystal structure of the Trypanosoma cruzi protein tyrosine phosphatase TcPTP1, a potential therapeutic target for Chagas' disease | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, TYROSINE SPECIFIC PROTEIN PHOSPHATASE | | Authors: | Lountos, G.T, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2012-06-22 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Structure of the Trypanosoma Cruzi Protein Tyrosine Phosphatase Tcptp1, a Potential Therapeutic Target for Chagas' Disease.
Mol.Biochem.Parasitol., 187, 2012
|
|
3I36
 
 | |
4BPC
 
 | |
3I7Z
 
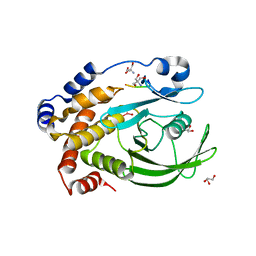 | | Protein Tyrosine Phosphatase 1B - Transition state analog for the first catalytic step | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EGFR receptor fragment, GLYCEROL, ... | | Authors: | Brandao, T.A.S, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2009-07-09 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into the reaction of protein-tyrosine phosphatase 1B: crystal structures for transition state analogs of both catalytic steps.
J.Biol.Chem., 285, 2010
|
|
3I80
 
 | | Protein Tyrosine Phosphatase 1B - Transition state analog for the second catalytic step | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, ... | | Authors: | Brandao, T.A.S, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2009-07-09 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into the reaction of protein-tyrosine phosphatase 1B: crystal structures for transition state analogs of both catalytic steps.
J.Biol.Chem., 285, 2010
|
|
3MOW
 
 | | Crystal structure of SHP2 in complex with a tautomycetin analog TTN D-1 | | Descriptor: | (2Z)-2-[(1R)-3-{[(1R,2S,3R,6S,7S,10S,12S,15E,17E)-18-carboxy-16-ethyl-3,7-dihydroxy-1,2,6,10,12-pentamethyl-5-oxooctade ca-15,17-dien-1-yl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Liu, S, Yu, Z, Yu, X. | | Deposit date: | 2010-04-23 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SHP2 in complex with a tautomycetin analog TTN D-1
To be Published
|
|
3M4U
 
 | |
8GVV
 
 | | PTPN21 PTP domain C1108S mutant | | Descriptor: | IODIDE ION, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8GWH
 
 | |
8JBN
 
 | | Vascular endothelial protein tyrosine phosphatase in complex with Cpd-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(1~{H}-indol-3-yl)-1,2-oxazole-3-carboxylic acid, ... | | Authors: | Orita, T, Furuzono, T, Doi, S, Adachi, T. | | Deposit date: | 2023-05-09 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fragment-Based Discovery of Novel VE-PTP Inhibitors Using Orthogonal Biophysical Techniques.
Biochemistry, 62, 2023
|
|
8JBY
 
 | | Vascular endothelial protein tyrosine phosphatase in complex with Cpd-2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-(hydroxymethyl)-5-(1-methylindol-3-yl)-1,2-oxazole-3-carboxylic acid, ... | | Authors: | Orita, T, Furuzono, T, Doi, S, Adachi, T. | | Deposit date: | 2023-05-10 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fragment-Based Discovery of Novel VE-PTP Inhibitors Using Orthogonal Biophysical Techniques.
Biochemistry, 62, 2023
|
|
