8S9P
 
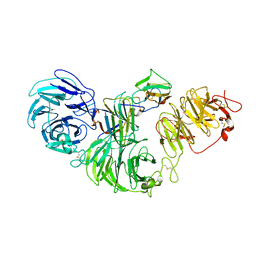 | | 1:1:1 agrin/LRP4/MuSK complex | | Descriptor: | Agrin, Low-density lipoprotein receptor-related protein 4, Muscle, ... | | Authors: | Xie, T, Xu, G.J, Liu, Y, Quade, B, Lin, W.C, Bai, X.C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-17 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the assembly of the agrin/LRP4/MuSK signaling complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7QE9
 
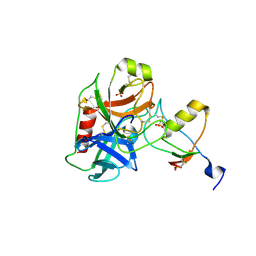 | | Human cationic trypsin (TRY1) complexed with serine protease inhibitor Kazal type 1 N34S (SPINK1 N34S) | | Descriptor: | SULFATE ION, Serine protease inhibitor Kazal-type 1, Trypsin-1 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin.
Int J Mol Sci, 23, 2022
|
|
7QE8
 
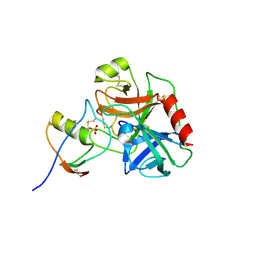 | | Human cationic trypsin (TRY1) complexed with serine protease inhibitor Kazal type 1 (SPINK1) | | Descriptor: | SULFATE ION, Serine protease inhibitor Kazal-type 1, Trypsin-1 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin.
Int J Mol Sci, 23, 2022
|
|
7E50
 
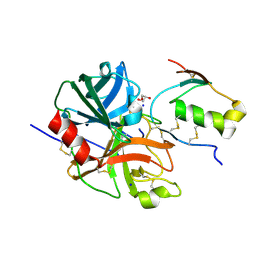 | | Crystal structure of human microplasmin in complex with kazal-type inhibitor AaTI | | Descriptor: | AAEL006007-PA, GLYCEROL, Plasminogen, ... | | Authors: | Varsha, A.W, Jobichen, C, Mok, Y.K. | | Deposit date: | 2021-02-16 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Aedes aegypti trypsin inhibitor in complex with mu-plasmin reveals role for scaffold stability in Kazal-type serine protease inhibitor.
Protein Sci., 31, 2022
|
|
6KBR
 
 | |
5YHN
 
 | | Solution structure of the LEKTI Domain 4 | | Descriptor: | cDNA FLJ60407, highly similar to Serine protease inhibitor Kazal-type 5 | | Authors: | Mok, Y.K, Ramesh, K. | | Deposit date: | 2017-09-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Homologous Lympho-Epithelial Kazal-type Inhibitor Domains Delay Blood Coagulation by Inhibiting Factor X and XI with Differential Specificity.
Structure, 26, 2018
|
|
2N71
 
 | | NMR structure of CmPI-II, a serin protease inhibitor isolated from mollusk Cenchitis muricatus | | Descriptor: | Protease inhibitor 2 | | Authors: | Cabrera-Munoz, A, Rojas, L, Alonso del Rivero Antigua, M, Pires, J. | | Deposit date: | 2015-09-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of CmPI-II, a non-classical Kazal protease inhibitor: Understanding its conformational dynamics and subtilisin A inhibition.
J.Struct.Biol., 206, 2019
|
|
5DAE
 
 | |
2N52
 
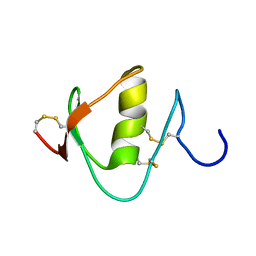 | |
2M5X
 
 | | Novel method of protein purification for structural research. Example of ultra high resolution structure of SPI-2 inhibitor by X-ray and NMR spectroscopy. | | Descriptor: | Silk protease inhibitor 2 | | Authors: | Lenarcic Zivkovic, M, Dvornyk, A, Kludkiewicz, B, Kopera, E, Zagorski-Ostoja, W, Grzelak, K, Zhukov, I, Bal, W. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
4HGU
 
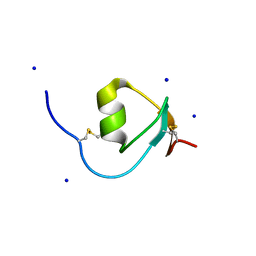 | | Crystal Structure of Galleria mellonella Silk Protease Inhibitor 2 | | Descriptor: | SODIUM ION, Silk protease inhibitor 2 | | Authors: | Krzywda, S, Jaskolski, M, Dvornyk, A, Kludkiewicz, B, Grzelak, K, Zagorski, W, Bal, W, Kopera, E. | | Deposit date: | 2012-10-08 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
2LEO
 
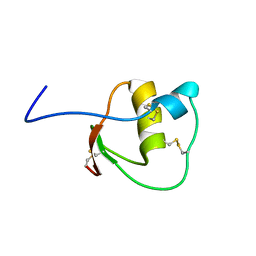 | |
2KMO
 
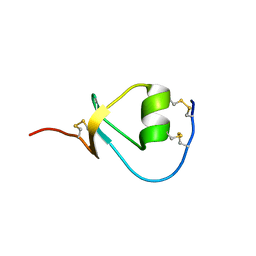 | |
2KMR
 
 | |
2KMP
 
 | |
2KMQ
 
 | |
2V53
 
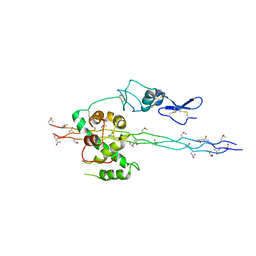 | | Crystal structure of a SPARC-collagen complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COLLAGEN ALPHA-1(III) CHAIN, ... | | Authors: | Hohenester, E, Sasaki, T, Giudici, C, Farndale, R.W, Bachinger, H.P. | | Deposit date: | 2008-10-01 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Sequence-Specific Collagen Recognition by Sparc.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2JXD
 
 | |
2GKR
 
 | | Crystal structure of the N-terminally truncated OMTKY3-del(1-5) | | Descriptor: | CHLORIDE ION, Ovomucoid | | Authors: | Lee, T.W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-04-03 | | Release date: | 2007-02-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Insights into the Non-additivity Effects in the Sequence-to-Reactivity Algorithm for Serine Peptidases and their Inhibitors.
J.Mol.Biol., 367, 2007
|
|
2GKV
 
 | | Crystal structure of the SGPB:P14'-Ala32 OMTKY3-del(1-5) complex | | Descriptor: | Ovomucoid, Streptogrisin B | | Authors: | Lee, T.W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-04-03 | | Release date: | 2007-02-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Non-additivity Effects in the Sequence-to-Reactivity Algorithm for Serine Peptidases and their Inhibitors.
J.Mol.Biol., 367, 2007
|
|
2GKT
 
 | |
2F3C
 
 | | Crystal structure of infestin 1, a Kazal-type serineprotease inhibitor, in complex with trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Campos, I.T.N, Tanaka, A.S, Barbosa, J.A.R.G. | | Deposit date: | 2005-11-20 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Kazal-type inhibitors infestins 1 and 4 differ in specificity but are similar in three-dimensional structure.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2NU0
 
 | | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I, and Tyr18I | | Descriptor: | Ovomucoid, Streptogrisin B, Protease B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I, and Tyr18I
To be Published
|
|
2NU1
 
 | | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I | | Descriptor: | Ovomucoid, Streptogrisin B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I
To be Published
|
|
2NU4
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Proteinase B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
