8SCB
 
 | | Terminating ribosome with SRI-41315 | | Descriptor: | (2S,4aS)-2-cyclobutyl-10-methyl-3-phenyl-2,10-dihydropyrimido[4,5-b]quinoline-4,5(3H,4aH)-dione, 18S_rRNA, 28S_rRNA, ... | | Authors: | Yip, M.C.J, Coelho, J.P.L, Oltion, K, Tauton, J, Shao, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and consequences of eRF1 glued to the ribosomal decoding center
To Be Published
|
|
8P09
 
 | | 48S late-stage initiation complex with non methylated mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Guca, E, Lima, L.H.F, Boissier, F, Hashem, Y. | | Deposit date: | 2023-05-09 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | N 6 -methyladenosine in 5' UTR does not promote translation initiation.
Mol.Cell, 84, 2024
|
|
8P03
 
 | | 48S late-stage initiation complex with m6A mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Guca, E, Lima, L.H.F, Boissier, F, Hashem, Y. | | Deposit date: | 2023-05-09 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | N 6 -methyladenosine in 5' UTR does not promote translation initiation.
Mol.Cell, 84, 2024
|
|
8OH9
 
 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Formate dehydrogenase-O, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
8OH5
 
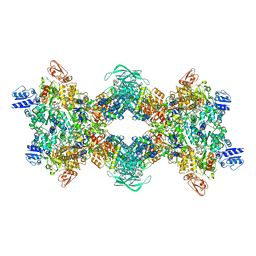 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 2) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
8CAS
 
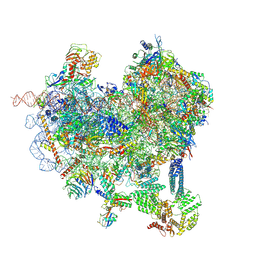 | | Cryo-EM structure of native Otu2-bound ubiquitinated 48S initiation complex (partial) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8CAH
 
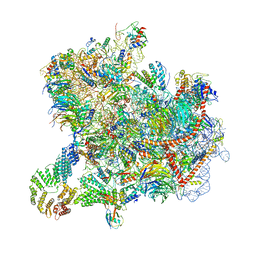 | | Cryo-EM structure of native Otu2-bound ubiquitinated 43S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
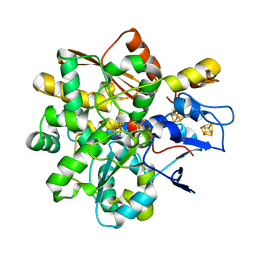
8BJ8
 
 | |
8BJ7
 
 | |
7ZCI
 
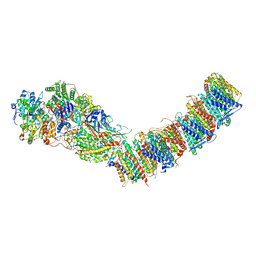 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-28 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZC5
 
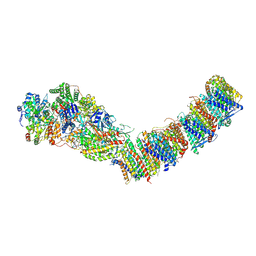 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z84
 
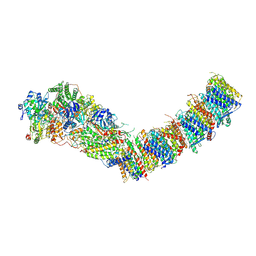 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z83
 
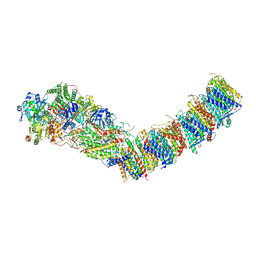 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z80
 
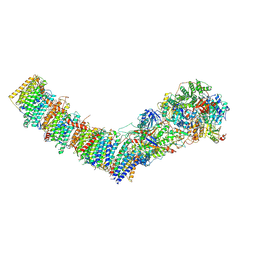 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7V
 
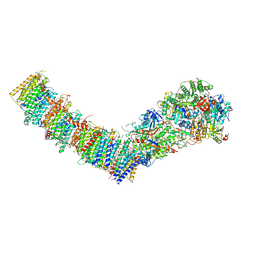 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7T
 
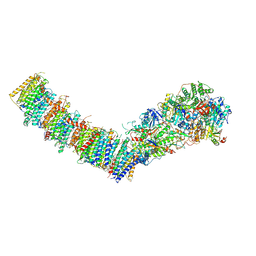 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7S
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7R
 
 | | Complex I from E. coli, LMNG-purified, Apo, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z0T
 
 | | Structure of the Escherichia coli formate hydrogenlyase complex (aerobic preparation, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARBONMONOXIDE-(DICYANO) IRON, FE (III) ION, ... | | Authors: | Steinhilper, R, Murphy, B.J. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the membrane-bound formate hydrogenlyase complex from Escherichia coli.
Nat Commun, 13, 2022
|
|
7Z0S
 
 | |
7T30
 
 | |
7T2R
 
 | |
7QV7
 
 | | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | | Descriptor: | Hydrogen dependent carbon dioxide reductase subunit FdhF, Hydrogen dependent carbon dioxide reductase subunit HycB3, Hydrogen dependent carbon dioxide reductase subunit HycB4, ... | | Authors: | Dietrich, H.M, Righetto, R.D, Kumar, A, Wietrzynski, W, Schuller, S.K, Trischler, R, Wagner, J, Schwarz, F.M, Engel, B.D, Mueller, V, Schuller, J.M. | | Deposit date: | 2022-01-19 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Nature, 607, 2022
|
|
7Q5Y
 
 | |
7PLM
 
 | | CryoEM reconstruction of pyruvate ferredoxin oxidoreductase (PFOR) in anaerobic conditions | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cherrier, M.V, Vernede, X, Fenel, D, Martin, L, Arragain, B, Neumann, E, Fontecilla Camps, J.C, Schoehn, G, Nicolet, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Oxygen-Sensitive Metalloprotein Structure Determination by Cryo-Electron Microscopy.
Biomolecules, 12, 2022
|
|
