5IBQ
 
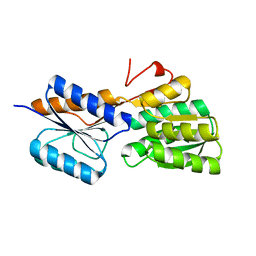 | | Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose | | Descriptor: | 3-C-(hydroxylmethyl)-alpha-D-erythrofuranose, CALCIUM ION, Probable ribose ABC transporter, ... | | Authors: | Vetting, M.W, Carter, M.S, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose
To be published
|
|
1VJW
 
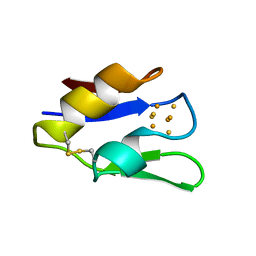 | | STRUCTURE OF OXIDOREDUCTASE (NADP+(A),FERREDOXIN(A)) | | Descriptor: | FERREDOXIN(A), IRON/SULFUR CLUSTER | | Authors: | Macedo-Ribeiro, S, Darimont, B, Sterner, R, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1996-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small structural changes account for the high thermostability of 1[4Fe-4S] ferredoxin from the hyperthermophilic bacterium Thermotoga maritima.
Structure, 4, 1996
|
|
1XER
 
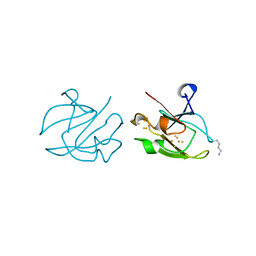 | | STRUCTURE OF FERREDOXIN | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, ZINC ION | | Authors: | Fujii, T, Hata, Y, Moriyama, H, Wakagi, T, Tanaka, N, Oshima, T. | | Deposit date: | 1996-08-28 | | Release date: | 1997-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding centre in thermoacidophilic archaeal ferredoxins.
Nat.Struct.Biol., 3, 1996
|
|
2ZVS
 
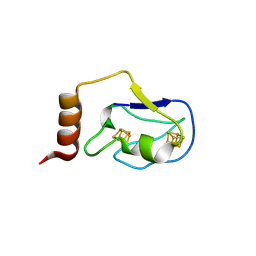 | | Crystal structure of the 2[4FE-4S] ferredoxin from escherichia coli | | Descriptor: | IRON/SULFUR CLUSTER, Uncharacterized ferredoxin-like protein yfhL | | Authors: | Giastas, P, Mavridis, M.I. | | Deposit date: | 2008-11-18 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insight into the protein and solvent contributions to the reduction potentials of [4Fe-4S]2+/+ clusters: crystal structures of the Allochromatium vinosum ferredoxin variants C57A and V13G and the homologous Escherichia coli ferredoxin
J.Biol.Inorg.Chem., 14, 2009
|
|
3EUN
 
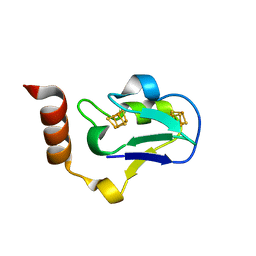 | |
3EXY
 
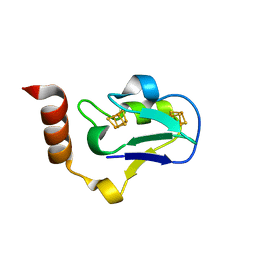 | |
2V2K
 
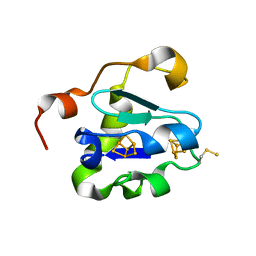 | | THE CRYSTAL STRUCTURE OF FDXA, A 7FE FERREDOXIN FROM MYCOBACTERIUM SMEGMATIS | | Descriptor: | ACETATE ION, FE3-S4 CLUSTER, FERREDOXIN | | Authors: | Ricagno, S, de Rosa, M, Aliverti, A, Zanetti, G, Bolognesi, M. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Fdxa, a 7Fe Ferredoxin from Mycobacterium Smegmatis.
Biochem.Biophys.Res.Commun., 360, 2007
|
|
1RGV
 
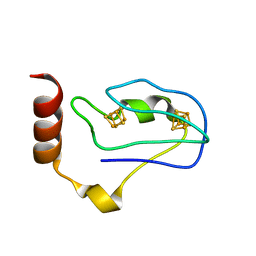 | | Crystal Structure of the Ferredoxin from Thauera aromatica | | Descriptor: | IRON/SULFUR CLUSTER, ferredoxin | | Authors: | Unciuleac, M, Boll, M, Warkentin, E, Ermler, U. | | Deposit date: | 2003-11-13 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallization of 4-hydroxybenzoyl-CoA reductase and the structure of its electron donor ferredoxin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ROF
 
 | | NMR STUDY OF 4FE-4S FERREDOXIN OF THERMATOGA MARITIMA | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Roesch, P, Sticht, H, Wildegger, G, Bentrop, D, Darimont, B, Sterner, R. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An NMR-derived model for the solution structure of oxidized Thermotoga maritima 1[Fe4-S4] ferredoxin.
Eur.J.Biochem., 237, 1996
|
|
1BC6
 
 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, 20 STRUCTURES | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-05 | | Release date: | 1998-06-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1BD6
 
 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1BLU
 
 | | STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CHROMATIUM VINOSUM | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Moulis, J.M. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the 2[4Fe-4S] ferredoxin from Chromatium vinosum: evolutionary and mechanistic inferences for [3/4Fe-4S] ferredoxins.
Protein Sci., 5, 1996
|
|
1BQX
 
 | | ARTIFICIAL FE8S8 FERREDOXIN: THE D13C VARIANT OF BACILLUS SCHLEGELII FE7S8 FERREDOXIN | | Descriptor: | IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN) | | Authors: | Aono, S, Bentrop, D, Bertini, I, Cosenza, G, Luchinat, C. | | Deposit date: | 1998-08-20 | | Release date: | 1998-08-26 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an artificial Fe8S8 ferredoxin: the D13C variant of Bacillus schlegelii Fe7S8 ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
1BWE
 
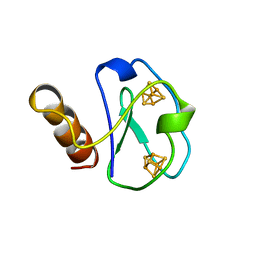 | | ARTIFICIAL FE8S8 FERREDOXIN: THE D13C VARIANT OF BACILLUS SCHLEGELII FE7S8 FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Cosenza, G, Luchinat, C. | | Deposit date: | 1998-09-23 | | Release date: | 1998-09-30 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an artificial Fe8S8 ferredoxin: the D13C variant of Bacillus schlegelii Fe7S8 ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
2FDN
 
 | | 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDI-URICI | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Meyer, J, Moulis, J.M. | | Deposit date: | 1997-10-01 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Atomic resolution (0.94 A) structure of Clostridium acidurici ferredoxin. Detailed geometry of [4Fe-4S] clusters in a protein.
Biochemistry, 36, 1997
|
|
1DUR
 
 | |
1FCA
 
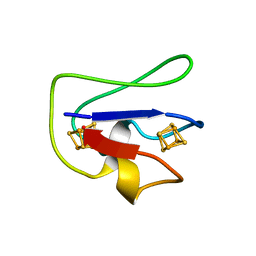 | |
1FDN
 
 | | REFINED CRYSTAL STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDURICI AT 1.84 ANGSTROMS RESOLUTION | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Duee, E, Fanchon, E, Vicat, J, Sieker, L.C, Meyer, J, Moulis, J-M. | | Deposit date: | 1994-03-31 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Refined crystal structure of the 2[4Fe-4S] ferredoxin from Clostridium acidurici at 1.84 A resolution.
J.Mol.Biol., 243, 1994
|
|
1H98
 
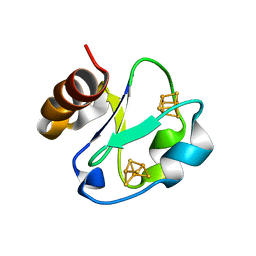 | | New Insights into Thermostability of Bacterial Ferredoxins: High Resolution Crystal Structure of the Seven-Iron Ferredoxin from Thermus thermophilus | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Macedo-Ribeiro, S, Martins, B.M, Pereira, P.J.B, Buse, G, Huber, R, Soulimane, T. | | Deposit date: | 2001-03-05 | | Release date: | 2001-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | New Insights Into the Thermostability of Bacterial Ferredoxins: High-Resolution Crystal Structure of the Seven-Iron Ferredoxin from Thermus Thermophilus
J.Biol.Inorg.Chem., 6, 2001
|
|
1D3W
 
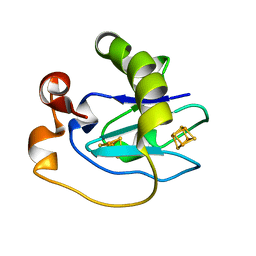 | | Crystal structure of ferredoxin 1 d15e mutant from azotobacter vinelandii at 1.7 angstrom resolution. | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER | | Authors: | Chen, K, Hirst, J, Camba, R, Bonagura, C.A, Stout, C.D, Burges, B.K, Armstrong, F.A. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomically defined mechanism for proton transfer to a buried redox centre in a protein.
Nature, 405, 2000
|
|
4HEA
 
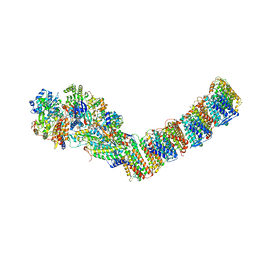 | | Crystal structure of the entire respiratory complex I from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Baradaran, R, Berrisford, J.M, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3027 Å) | | Cite: | Crystal structure of the entire respiratory complex I.
Nature, 494, 2013
|
|
5C4I
 
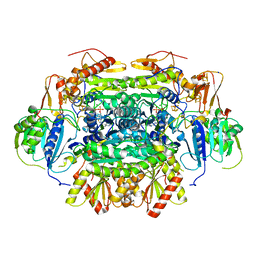 | | Structure of an Oxalate Oxidoreductase | | Descriptor: | IRON/SULFUR CLUSTER, MAGNESIUM ION, Oxalate oxidoreductase subunit alpha, ... | | Authors: | Gibson, M.I, Brignole, E.J, Pierce, E, Can, M, Ragsdale, S.W, Drennan, C.L. | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | The Structure of an Oxalate Oxidoreductase Provides Insight into Microbial 2-Oxoacid Metabolism.
Biochemistry, 54, 2015
|
|
3MMA
 
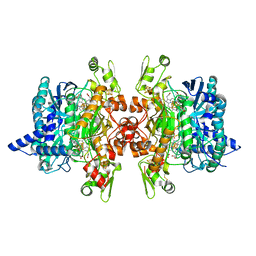 | | Dissimilatory sulfite reductase phosphate complex | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
6Q8W
 
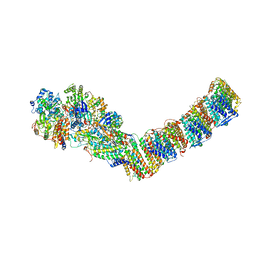 | | Respiratory complex I from Thermus thermophilus with bound Aureothin. | | Descriptor: | Aureothin, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-12-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
3MMB
 
 | | Dissimilatory sulfite reductase in complex with the endproduct sulfide | | Descriptor: | HYDROSULFURIC ACID, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
