2CPD
 
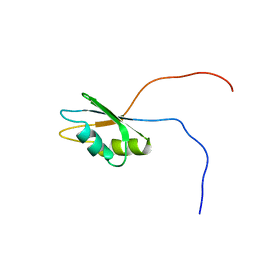 | | Solution structure of the RNA recognition motif of human APOBEC-1 complementation factor, ACF | | Descriptor: | APOBEC-1 stimulating protein | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA recognition motif of human APOBEC-1 complementation factor, ACF
To be Published
|
|
1YNE
 
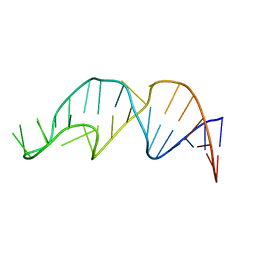 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | APOLIPOPROTEIN B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YNG
 
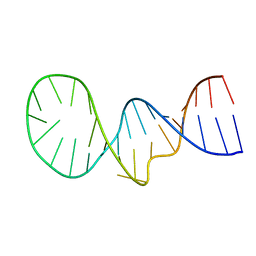 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YNC
 
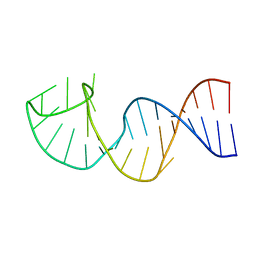 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YLG
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
2G5T
 
 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) complexed with cyanopyrrolidine (C5-pro-pro) inhibitor 21ag | | Descriptor: | 3-{[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHOXY}-4-CHLOROBENZOIC ACID, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Fry, E.H, Lake, M.R, Solomon, L.R, Pei, Z, Li, X. | | Deposit date: | 2006-02-23 | | Release date: | 2006-07-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery, structure-activity relationship, and pharmacological evaluation of (5-substituted-pyrrolidinyl-2-carbonyl)-2-cyanopyrrolidines as potent dipeptidyl peptidase IV inhibitors.
J.Med.Chem., 49, 2006
|
|
6ND5
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with chloramphenicol and bound to mRNA and A-, P-, and E-site tRNAs at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
4V9Q
 
 | | Crystal Structure of Blasticidin S Bound to Thermus Thermophilus 70S Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Ling, C, Ermolenko, D.N, Korostelev, A.A. | | Deposit date: | 2013-06-12 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V6F
 
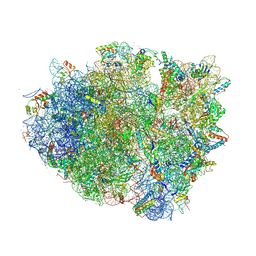 | | Elongation complex of the 70S ribosome with three tRNAs and mRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 23S RRNA, ... | | Authors: | Jenner, L.B, Yusupova, G, Yusupov, M. | | Deposit date: | 2009-07-09 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural aspects of messenger RNA reading frame maintenance by the ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3PCE
 
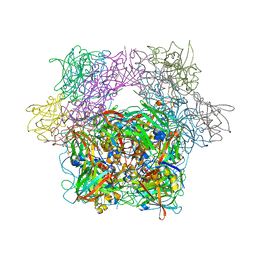 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-HYDROXYPHENYLACETATE | | Descriptor: | 3-HYDROXYPHENYLACETATE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Elango, N, Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-04-29 | | Release date: | 1998-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCC
 
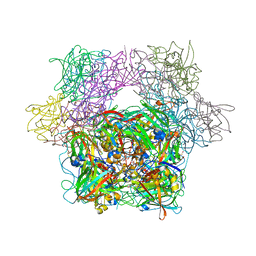 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 4-HYDROXYBENZOATE | | Descriptor: | BETA-MERCAPTOETHANOL, FE (III) ION, P-HYDROXYBENZOIC ACID, ... | | Authors: | Elango, N, Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-04-29 | | Release date: | 1998-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCF
 
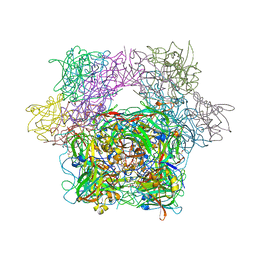 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-FLURO-4-HYDROXYBENZOATE | | Descriptor: | 3-FLUORO-4-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-06-27 | | Release date: | 1998-01-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCG
 
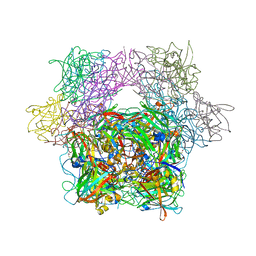 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH THE INHIBITOR 4-HYDROXYPHENYLACETATE | | Descriptor: | 4-HYDROXYPHENYLACETATE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Elango, N, Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-04-29 | | Release date: | 1998-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCB
 
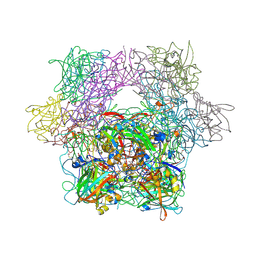 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-HYDROXYBENZOATE | | Descriptor: | 3-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Elango, N, Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-04-25 | | Release date: | 1998-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCD
 
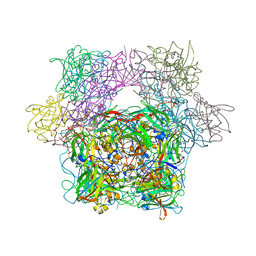 | | PROTOCATECHUATE 3,4-DIOXYGENASE Y447H MUTANT | | Descriptor: | BETA-MERCAPTOETHANOL, CARBONATE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The axial tyrosinate Fe3+ ligand in protocatechuate 3,4-dioxygenase influences substrate binding and product release: evidence for new reaction cycle intermediates.
Biochemistry, 37, 1998
|
|
3PCI
 
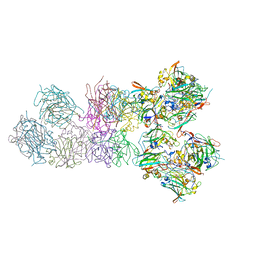 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-IODO-4-HYDROXYBENZOATE | | Descriptor: | 3-IODO-4-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCH
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-CHLORO-4-HYDROXYBENZOATE | | Descriptor: | 3-CHLORO-4-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-01 | | Release date: | 1998-01-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCM
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 6-HYDROXYNICOTINIC ACID N-OXIDE AND CYANIDE | | Descriptor: | 6-HYDROXYISONICOTINIC ACID N-OXIDE, CYANIDE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCA
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3,4-DIHYDROXYBENZOATE | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCK
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 6-HYDROXYNICOTINIC ACID N-OXIDE | | Descriptor: | 6-HYDROXYISONICOTINIC ACID N-OXIDE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCL
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 2-HYDROXYISONICOTINIC ACID N-OXIDE AND CYANIDE | | Descriptor: | 2-HYDROXYISONICOTINIC ACID N-OXIDE, CYANIDE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCJ
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 2-HYDROXYISONICOTINIC ACID N-OXIDE | | Descriptor: | 2-HYDROXYISONICOTINIC ACID N-OXIDE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCN
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3,4-DIHYDROXYPHENYLACETATE | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-08-19 | | Release date: | 1998-02-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and resonance Raman studies of protocatechuate 3,4-dioxygenase complexed with 3,4-dihydroxyphenylacetate.
Biochemistry, 36, 1997
|
|
2BYM
 
 | | Histone fold heterodimer of the Chromatin Accessibility Complex | | Descriptor: | CADMIUM ION, CHRAC-14, CHRAC-16 | | Authors: | Fernandez-Tornero, C, Hartlepp, K.F, Grune, T, Eberharter, A, Becker, P.B, Muller, C.W. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Histone Fold Subunits of Drosophila Chrac Facilitate Nucleosome Sliding Through Dynamic DNA Interactions.
Mol.Cell.Biol., 25, 2005
|
|
