3MUV
 
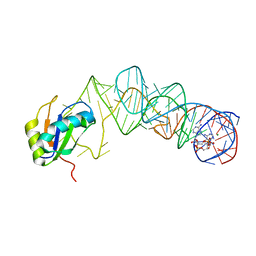 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
7QZP
 
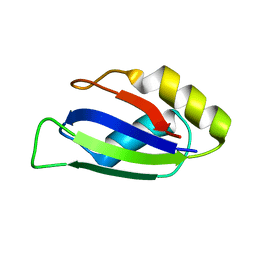 | | Identification and characterization of an RRM-containing, ELAV-like, RNA binding protein in Acinetobacter Baumannii | | Descriptor: | Hypothetical RNA binding protein from Acinetobacter baumannii | | Authors: | Ciani, C, Perez-Rafols, A, Bonomo, I, Micaelli, M, Esposito, A, Zucal, C, Belli, R, D'Agostino, V.G, Bianconi, I, Calderone, V, Cerofolini, L, Fragai, M, Provenzani, A. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification and Characterization of an RRM-Containing, RNA Binding Protein in Acinetobacter baumannii .
Biomolecules, 12, 2022
|
|
7QR3
 
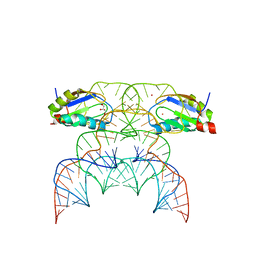 | | Chimpanzee CPEB3 HDV-like ribozyme | | Descriptor: | GLYCEROL, POTASSIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Przytula-Mally, A.I, Engilberge, S, Johannsen, S, Olieric, V, Masquida, B, Sigel, R.K.O. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Anticodon-like loop-mediated dimerization in the crystal structures of HdV-like CPEB3 ribozymes
Biorxiv, 2022
|
|
7QR4
 
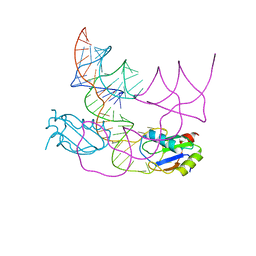 | | human CPEB3 HDV-like ribozyme | | Descriptor: | RNA CPEB3 ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Przytula-Mally, A.I, Engilberge, S, Johannsen, S, Olieric, V, Masquida, B, Sigel, R.K.O. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Anticodon-like loop-mediated dimerization in the crystal structures of HdV-like CPEB3 ribozymes
Biorxiv, 2022
|
|
3NMR
 
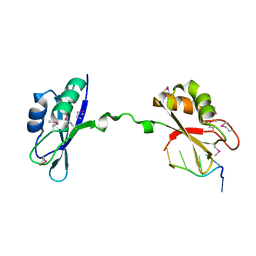 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3NNC
 
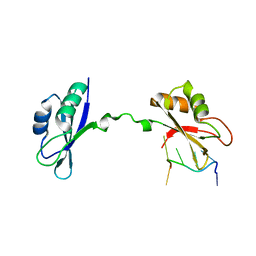 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*UP*GP*UP*GP*UP*G)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2005 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3NNA
 
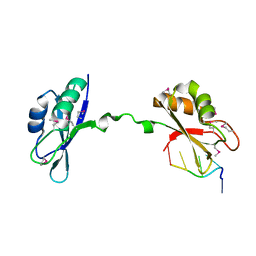 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3NS6
 
 | |
3NNH
 
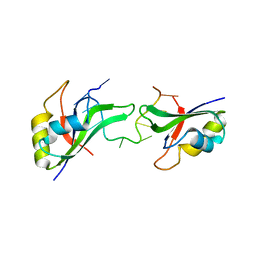 | | Crystal Structure of the CUGBP1 RRM1 with GUUGUUUUGUUU RNA | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7501 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3NS5
 
 | |
7S3B
 
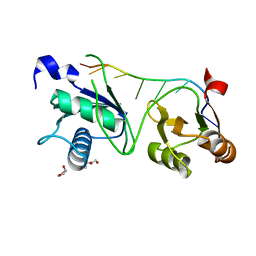 | |
7S3A
 
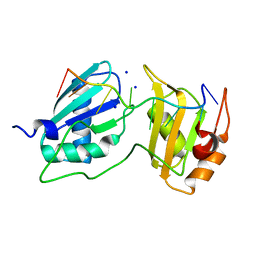 | |
7S3C
 
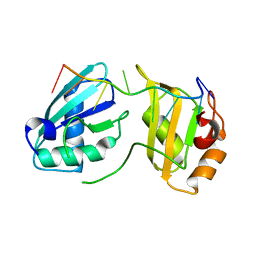 | |
7SN6
 
 | | U2AF65 UHM BOUND TO SF3B155 ULM5 | | Descriptor: | ISOPROPYL ALCOHOL, Splicing factor 3B subunit 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2021-10-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A UHM-ULM interface with unusual structural features contributes to U2AF2 and SF3B1 association for pre-mRNA splicing.
J.Biol.Chem., 298, 2022
|
|
3P49
 
 | | Crystal Structure of a Glycine Riboswitch from Fusobacterium nucleatum | | Descriptor: | GLYCINE, GLYCINE RIBOSWITCH, MAGNESIUM ION, ... | | Authors: | Butler, E.B, Wang, J, Xiong, Y, Strobel, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of cooperative ligand binding by the glycine riboswitch.
Chem.Biol., 18, 2011
|
|
3R1H
 
 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Ca2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', CALCIUM ION, Class I ligase ribozyme, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
5X3Z
 
 | | Solution structure of musashi1 RBD2 in complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | Descriptor: | RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5ZUH
 
 | |
6C8U
 
 | | Solution structure of Musashi2 RRM1 | | Descriptor: | RNA-binding protein Musashi homolog 2 | | Authors: | Xing, M, Lan, L, Douglas, J.T, Gao, P, Hanzlik, R.P, Xu, L. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2022-03-23 | | Method: | SOLUTION NMR | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 2019
|
|
6CMN
 
 | | Co-Crystal Structure of HIV-1 TAR Bound to Lab-Evolved RRM TBP6.7 | | Descriptor: | TAR-Binding Protein 6.7, Trans-Activation Response RNA Element | | Authors: | Belashov, I.A, Wedekind, J.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structure of HIV TAR in complex with a Lab-Evolved RRM provides insight into duplex RNA recognition and synthesis of a constrained peptide that impairs transcription.
Nucleic Acids Res., 46, 2018
|
|
6XH3
 
 | |
6XH2
 
 | |
6XLX
 
 | |
7VSJ
 
 | |
