4P9O
 
 | |
4PA7
 
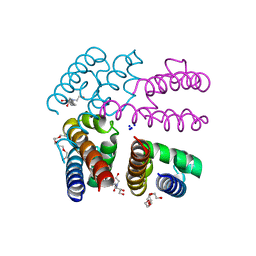 | | Structure of NavMS pore and C-terminal domain crystallised in presence of channel blocking compound | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7PGI
 
 | | NaVAb1p (bicelles) | | Descriptor: | ACETATE ION, Ion transport protein, MAGNESIUM ION, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.638 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGF
 
 | |
7PG8
 
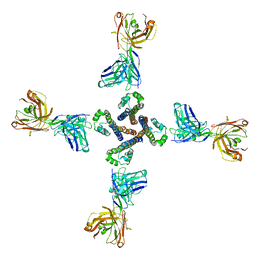 | | NaV_Ae1/Sp1CTD_pore-ANT05 complex | | Descriptor: | ANT05 H12 fab fragment, heavy chain, light chain, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGB
 
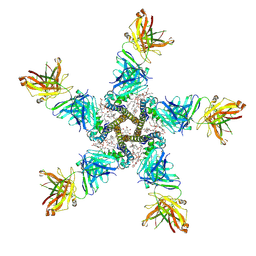 | | NaV_Ae1/Sp1CTD_pore-SAT09 complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-NITROBENZOIC ACID, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGG
 
 | | NaVAb1p detergent (DM) | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, Ion transport protein | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGH
 
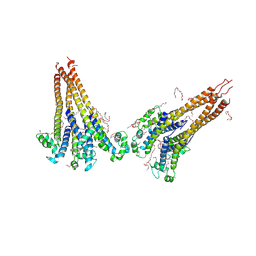 | | NaVAe1/Sp1CTDp (DDM) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DODECAETHYLENE GLYCOL, DODECANE, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (4.194 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5TUA
 
 | | structure of a Na+-selective mutant of two-pore channel from Arabidopsis thaliana AtTPC1 | | Descriptor: | BARIUM ION, CALCIUM ION, SODIUM ION, ... | | Authors: | Guo, J, Zeng, W, Jiang, Y. | | Deposit date: | 2016-11-05 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tuning the ion selectivity of two-pore channels.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VB8
 
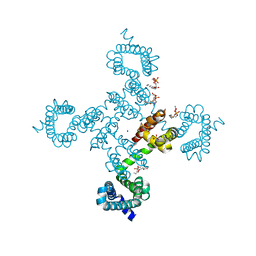 | |
5VB2
 
 | |
8H9X
 
 | |
8H9O
 
 | |
8H9W
 
 | |
8H9Y
 
 | |
8HA2
 
 | |
8HA1
 
 | |
6JUH
 
 | | structure of CavAb in complex with efonidipine | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[phenyl-(phenylmethyl)amino]ethyl (4~{R})-5-(5,5-dimethyl-2-oxidanylidene-1,3,2$l^{5}-dioxaphosphinan-2-yl)-2,6-dimethyl-4-(3-nitrophenyl)-1,4-dihydropyridine-3-carboxylate, ... | | Authors: | Tang, L, Xu, F. | | Deposit date: | 2019-04-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for efonidipine block of a voltage-gated Ca2+channel.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
6KE5
 
 | | Structure of CavAb in complex with Diltiazem and Amlodipine | | Descriptor: | CALCIUM ION, Ion transport protein, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Tang, L. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural Basis for Diltiazem Block of a Voltage-Gated Ca2+Channel.
Mol.Pharmacol., 96, 2019
|
|
6KEB
 
 | |
6KZP
 
 | | calcium channel-ligand | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N. | | Deposit date: | 2019-09-25 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Nature, 576, 2019
|
|
6KZO
 
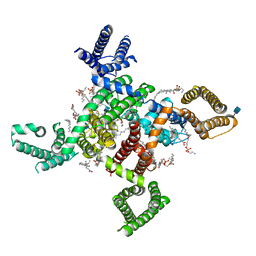 | | membrane protein | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N. | | Deposit date: | 2019-09-25 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Nature, 576, 2019
|
|
6MVW
 
 | | NavAb voltage-gated sodium channel, I217C/F203W | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein, PHOSPHATE ION | | Authors: | Lenaeus, M.J, Catterall, W.A. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Fenestrations control resting-state block of a voltage-gated sodium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6MIE
 
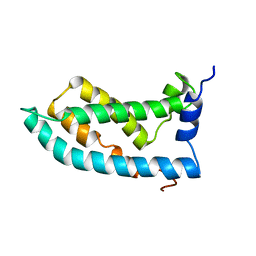 | | Solution NMR structure of the KCNQ1 voltage-sensing domain | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Taylor, K.C, Kuenze, G, Smith, J.A, Meiler, J, McFeeters, R.L, Sanders, C.R. | | Deposit date: | 2018-09-19 | | Release date: | 2020-03-04 | | Last modified: | 2020-03-11 | | Method: | SOLUTION NMR | | Cite: | Structure and physiological function of the human KCNQ1 channel voltage sensor intermediate state.
Elife, 9, 2020
|
|
6MWB
 
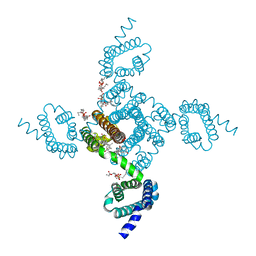 | | NavAb Voltage-gated Sodium Channel, residues 1-239 with mutation T206A | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein, ... | | Authors: | Lenaeus, M.J, Catterall, W.A. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular dissection of multiphase inactivation of the bacterial sodium channel NaVAb.
J. Gen. Physiol., 151, 2019
|
|
