3G8T
 
 | | Crystal structure of the G33A mutant Bacillus anthracis glmS ribozyme bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
1D9A
 
 | | SOLUTION STRUCTURE OF THE SECOND RNA-BINDING DOMAIN (RBD2) OF HU ANTIGEN C (HUC) | | Descriptor: | HU ANTIGEN C | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-07 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C.
Nucleic Acids Res., 28, 2000
|
|
1CVJ
 
 | | X-RAY CRYSTAL STRUCTURE OF THE POLY(A)-BINDING PROTEIN IN COMPLEX WITH POLYADENYLATE RNA | | Descriptor: | 5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', ADENOSINE-5'-MONOPHOSPHATE, POLYADENYLATE BINDING PROTEIN 1 | | Authors: | Deo, R.C, Bonanno, J.B, Sonenberg, N, Burley, S.K. | | Deposit date: | 1999-08-23 | | Release date: | 1999-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of polyadenylate RNA by the poly(A)-binding protein.
Cell(Cambridge,Mass.), 98, 1999
|
|
3G9C
 
 | | Crystal structure of the product Bacillus anthracis glmS ribozyme | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-13 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G96
 
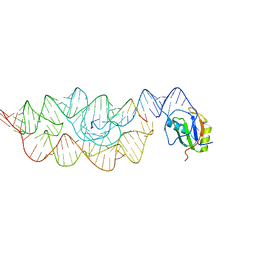 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to MaN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
1D8Z
 
 | | SOLUTION STRUCTURE OF THE FIRST RNA-BINDING DOMAIN (RBD1) OF HU ANTIGEN C (HUC) | | Descriptor: | HU ANTIGEN C | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-07 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C.
Nucleic Acids Res., 28, 2000
|
|
3B4D
 
 | | Crystal Structure of Human PABPN1 RRM | | Descriptor: | Polyadenylate-binding protein 2 | | Authors: | Ge, H, Tong, S, Teng, M, Niu, L. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure and possible dimerization of the single RRM of human PABPN1
Proteins, 71, 2008
|
|
3B4M
 
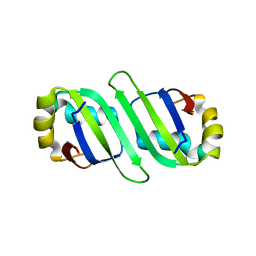 | | Crystal Structure of Human PABPN1 RRM | | Descriptor: | Polyadenylate-binding protein 2 | | Authors: | Ge, H, Zhou, D, Teng, M, Niu, L. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure and possible dimerization of the single RRM of human PABPN1
Proteins, 71, 2008
|
|
1DZ5
 
 | | The NMR structure of the 38KDa U1A protein-PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein | | Descriptor: | PIE, RNA (5'-R(*GP*AP*GP*AP*CP*AP*UP*UP*GP*CP*AP*CP*CP* CP*GP*GP*AP*GP*UP*CP*UP*C)-3'), U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Varani, L, Gunderson, S.I, Mattaj, I.W, Kay, L.E, Neuhaus, D, Varani, G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-03-29 | | Last modified: | 2013-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the 38kDa U1A Protein-Pie RNA Complex Reveals the Basis of Cooperativity in Regulation of Polyadenylation by Human U1A Protein
Nat.Struct.Biol., 7, 2000
|
|
3BO2
 
 | |
1DRZ
 
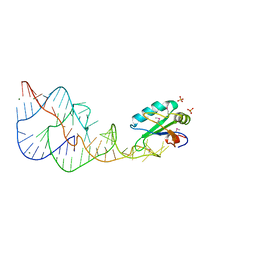 | | U1A SPLICEOSOMAL PROTEIN/HEPATITIS DELTA VIRUS GENOMIC RIBOZYME COMPLEX | | Descriptor: | MAGNESIUM ION, PROTEIN (U1 SMALL RIBONUCLEOPROTEIN A), RNA (HEPATITIS DELTA VIRUS GENOMIC RIBOZYME), ... | | Authors: | Ferre-D'Amare, A.R, Zhou, K, Doudna, J.A. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-16 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a hepatitis delta virus ribozyme.
Nature, 395, 1998
|
|
3BS9
 
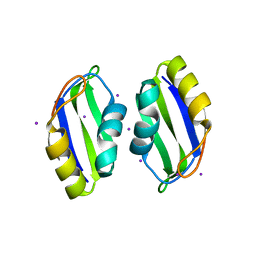 | | X-ray structure of human TIA-1 RRM2 | | Descriptor: | IODIDE ION, Nucleolysin TIA-1 isoform p40 | | Authors: | Kumar, A.O, Kielkopf, C.L. | | Deposit date: | 2007-12-22 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the central RNA recognition motif of human TIA-1 at 1.95A resolution.
Biochem.Biophys.Res.Commun., 367, 2008
|
|
3HHN
 
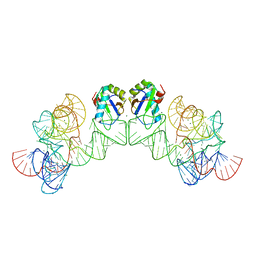 | | Crystal structure of class I ligase ribozyme self-ligation product, in complex with U1A RBD | | Descriptor: | Class I ligase ribozyme, self-ligation product, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Grant, R.A, Bagby, S.C, Bartel, D.P. | | Deposit date: | 2009-05-15 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.987 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
3HI9
 
 | |
5D77
 
 | | Structure of Mip6 RRM3 Domain | | Descriptor: | CITRIC ACID, NITRATE ION, RNA-binding protein MIP6, ... | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2015-08-13 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Mip6 RRM3 domain at 1.3
To Be Published
|
|
5CYJ
 
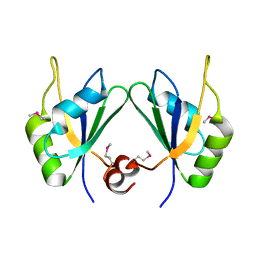 | | X-ray structure of human RBPMS | | Descriptor: | RNA-binding protein with multiple splicing | | Authors: | Teplova, M, Farazi, T.A, Patel, D.J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-09-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS.
Q. Rev. Biophys., 49, 2016
|
|
5DDP
 
 | | L-glutamine riboswitch bound with L-glutamine | | Descriptor: | GLUTAMINE, MAGNESIUM ION, RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDO
 
 | |
3BO3
 
 | |
3BO4
 
 | | A relaxed active site following exon ligation by a group I intron | | Descriptor: | DNA/RNA (5'-R(*AP*AP*GP*CP*CP*AP*CP*AP*CP*AP*AP*AP*CP*CP*A)-D(P*DG)-3'), DNA/RNA (5'-R(*CP*A)-D(P*DU)-R(P*AP*CP*GP*GP*CP*C)-3'), Group I intron P9, ... | | Authors: | Lipchock, S.V, Strobel, S.A. | | Deposit date: | 2007-12-17 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | A relaxed active site after exon ligation by the group I intron
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5DET
 
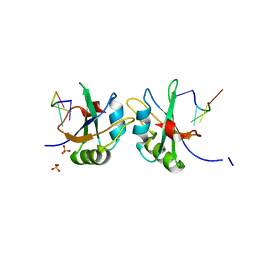 | | X-ray structure of human RBPMS in complex with the RNA | | Descriptor: | RNA (5'-R(*UP*CP*AP*C)-3'), RNA (5'-R(P*UP*CP*AP*CP*U)-3'), RNA-binding protein with multiple splicing, ... | | Authors: | Teplova, M, Farazi, T.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS.
Q. Rev. Biophys., 49, 2016
|
|
5D78
 
 | |
5DDQ
 
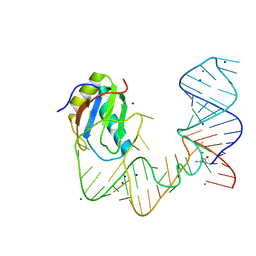 | | L-glutamine riboswitch bound with L-glutamine soaked with Mn2+ | | Descriptor: | GLUTAMINE, L-glutamine riboswitch RNA (61-MER), MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDR
 
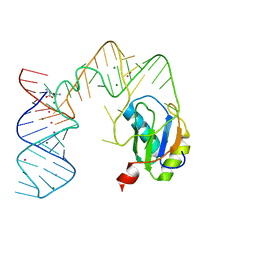 | | L-glutamine riboswitch bound with L-glutamine soaked with Cs+ | | Descriptor: | CESIUM ION, GLUTAMINE, L-glutamine riboswitch RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
3IRW
 
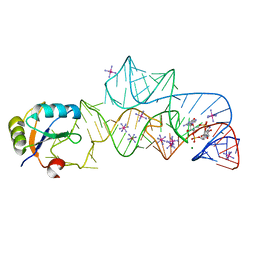 | | Structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Smith, K.D. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
