8U93
 
 | |
8U92
 
 | |
8UR1
 
 | |
6U01
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84D mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, L, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
4UR7
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with pyruvate | | Descriptor: | FORMIC ACID, GLYCEROL, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
6TZU
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, C, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
1YXD
 
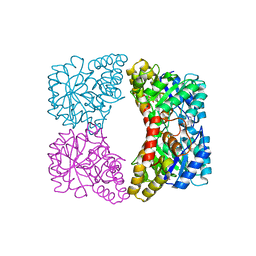 | | Structure of E. coli dihydrodipicolinate synthase bound with allosteric inhibitor (S)-lysine to 2.0 A | | Descriptor: | CHLORIDE ION, LYSINE, POTASSIUM ION, ... | | Authors: | Dobson, R.C.J, Griffin, M.D.W, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-02-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of native and (S)-lysine-bound dihydrodipicolinate synthase from Escherichia coli with improved resolution show new features of biological significance.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YXC
 
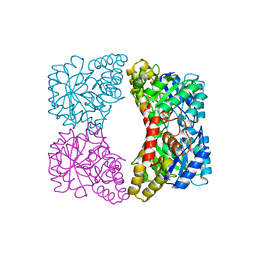 | | Structure of E. coli dihydrodipicolinate synthase to 1.9 A | | Descriptor: | CHLORIDE ION, POTASSIUM ION, dihydrodipicolinate synthase | | Authors: | Dobson, R.C.J, Griffin, M.D.W, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-02-20 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of native and (S)-lysine-bound dihydrodipicolinate synthase from Escherichia coli with improved resolution show new features of biological significance.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4UUI
 
 | |
4UR8
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with 2-oxoadipic acid | | Descriptor: | 2-OXOADIPIC ACID, FORMIC ACID, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
4UXD
 
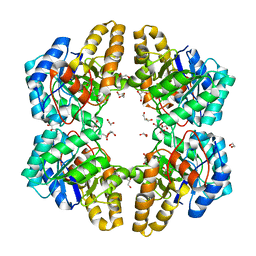 | | 2-keto 3-deoxygluconate aldolase from Picrophilus torridus | | Descriptor: | 1,2-ETHANEDIOL, 2-DEHYDRO-3-DEOXY-D-GLUCONATE/2-DEHYDRO-3-DEOXY-PHOSPHOGLUCONATE ALDOLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Priftis, A, Zaitsev, V, Reher, M, Johnsen, U, Danson, M.J, Taylor, G.L, Schoenheit, P, Crennell, S.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the Substrate Specificity of Archaeal Entner-Doudoroff Aldolases: The Structures of Picrophilus torridus 2-Keto-3-deoxygluconate Aldolase and Sulfolobus solfataricus 2-Keto-3-deoxy-6-phosphogluconate Aldolase in Complex with 2-Keto-3-deoxy-6-phosphogluconate.
Biochemistry, 57, 2018
|
|
4WOQ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric | | Descriptor: | 2-KETOBUTYRIC ACID, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-16 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric
to be published
|
|
4WOZ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine
to be published
|
|
2A6L
 
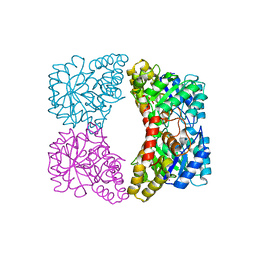 | | Dihydrodipicolinate synthase (E. coli)- mutant R138H | | Descriptor: | Dihydrodipicolinate synthase, POTASSIUM ION | | Authors: | Dobson, R.C, Devenish, S.R, Turner, L.A, Clifford, V.R, Pearce, F.G, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Role of Arginine 138 in the Catalysis and Regulation of Escherichia coli Dihydrodipicolinate Synthase.
Biochemistry, 44, 2005
|
|
2A6N
 
 | | Dihydrodipicolinate synthase (E. coli)- mutant R138A | | Descriptor: | Dihydrodipicolinate synthase, POTASSIUM ION | | Authors: | Dobson, R.C, Devenish, S.R, Turner, L.A, Clifford, V.R, Pearce, F.G, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Role of Arginine 138 in the Catalysis and Regulation of Escherichia coli Dihydrodipicolinate Synthase.
Biochemistry, 44, 2005
|
|
2ATS
 
 | | Dihydrodipicolinate synthase co-crystallised with (S)-lysine | | Descriptor: | CHLORIDE ION, D-LYSINE, POTASSIUM ION, ... | | Authors: | Devenish, S.R.A, Dobson, R.C.J, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-08-26 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The co-crystallisation of (S)-lysine-bound dihydrodipicolinate synthase from E. coli indicates that domain movements are not responsible for (S)-lysine inhibition
To be published
|
|
6VVI
 
 | |
6VVH
 
 | |
6XGS
 
 | |
2EHH
 
 | |
4HNN
 
 | |
1W3T
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with D-KDGal, D-Glyceraldehyde and pyruvate | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, 3-DEOXY-D-LYXO-HEXONIC ACID, D-Glyceraldehyde, ... | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-19 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
1W37
 
 | | 2-keto-3-deoxygluconate(KDG) aldolase of Sulfolobus solfataricus | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, GLYCEROL, SODIUM ION | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-02 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
1W3N
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with D-KDG | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, 3-DEOXY-D-ARABINO-HEXONIC ACID, GLYCEROL | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-17 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
1W3I
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with pyruvate | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, GLYCEROL, PYRUVIC ACID | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-15 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
