2GWM
 
 | |
2WN6
 
 | | Structural Basis for Substrate Recognition in the Enzymatic Component of ADP-ribosyltransferase Toxin CDTa from Clostridium difficile | | Descriptor: | ADP-RIBOSYLTRANSFERASE ENZYMATIC COMPONENT, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundriyal, A, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2009-07-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for Substrate Recognition in the Enzymatic Component of Adp-Ribosyltransferase Toxin Cdta from Clostridium Difficile.
J.Biol.Chem., 284, 2009
|
|
2WN7
 
 | | Structural Basis for Substrate Recognition in the Enzymatic Component of ADP-ribosyltransferase Toxin CDTa from Clostridium difficile | | Descriptor: | ADP-RIBOSYLTRANSFERASE ENZYMATIC COMPONENT, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sundriyal, A, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2009-07-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Substrate Recognition in the Enzymatic Component of Adp-Ribosyltransferase Toxin Cdta from Clostridium Difficile.
J.Biol.Chem., 284, 2009
|
|
2WN8
 
 | |
2C8B
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form II) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8G
 
 | | Structure of the PN loop Q182A mutant C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Properties of Wild-Type and Two Artt Motif Mutants Clostridium Botulinum C3 Exoenzyme Isoform 1 in Different Substrate Complexed States and Crystal Forms.
To be Published
|
|
2J3V
 
 | | Crystal structure of the enzymatic component C2-I of the C2-toxin from Clostridium botulinum at pH 3.0 | | Descriptor: | C2 TOXIN COMPONENT I, GLYCEROL, SULFATE ION | | Authors: | Schleberger, C, Hochmann, H, Barth, H, Aktories, K, Schulz, G.E. | | Deposit date: | 2006-08-23 | | Release date: | 2006-10-11 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure and Action of the Binary C2 Toxin from Clostridium Botulinum.
J.Mol.Biol., 364, 2006
|
|
2J3X
 
 | | Crystal structure of the enzymatic component C2-I of the C2-toxin from Clostridium botulinum at pH 3.0 (mut-S361R) | | Descriptor: | C2 TOXIN COMPONENT I, GLYCEROL, SULFATE ION | | Authors: | Schleberger, C, Hochmann, H, Barth, H, Aktories, K, Schulz, G.E. | | Deposit date: | 2006-08-23 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Action of the Binary C2 Toxin from Clostridium Botulinum.
J.Mol.Biol., 364, 2006
|
|
2J3Z
 
 | | Crystal structure of the enzymatic component C2-I of the C2-toxin from Clostridium botulinum at pH 6.1 | | Descriptor: | C2 TOXIN COMPONENT I, COBALT (II) ION, GLYCEROL, ... | | Authors: | Schleberger, C, Hochmann, H, Barth, H, Aktories, K, Schulz, G.E. | | Deposit date: | 2006-08-23 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Action of the Binary C2 Toxin from Clostridium Botulinum.
J.Mol.Biol., 364, 2006
|
|
4Y1W
 
 | |
4XZJ
 
 | | Crystal structure of ADP-ribosyltransferase Vis in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative NAD(+)--arginine ADP-ribosyltransferase Vis | | Authors: | Pfoh, R, Ravulapalli, R, Merrill, A.R, Pai, E.F. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of Vis Toxin, a Novel ADP-Ribosyltransferase from Vibrio splendidus.
Biochemistry, 54, 2015
|
|
4XZK
 
 | |
4YC0
 
 | |
1OJQ
 
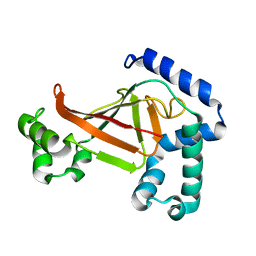 | | The crystal structure of C3stau2 from S. aureus | | Descriptor: | ADP-RIBOSYLTRANSFERASE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-15 | | Release date: | 2003-08-28 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
1OJZ
 
 | | The crystal structure of C3stau2 from S. aureus with NAD | | Descriptor: | ADP-RIBOSYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-16 | | Release date: | 2003-08-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
1QS2
 
 | | CRYSTAL STRUCTURE OF VIP2 WITH NAD | | Descriptor: | ADP-RIBOSYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Han, S, Craig, J.A, Putnam, C.D, Carozzi, N.B, Tainer, J.A. | | Deposit date: | 1999-06-25 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex.
Nat.Struct.Biol., 6, 1999
|
|
1QS1
 
 | | CRYSTAL STRUCTURE OF VEGETATIVE INSECTICIDAL PROTEIN2 (VIP2) | | Descriptor: | ADP-RIBOSYLTRANSFERASE | | Authors: | Han, S, Craig, J.A, Putnam, C.D, Carozzi, N.B, Tainer, J.A. | | Deposit date: | 1999-06-25 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex.
Nat.Struct.Biol., 6, 1999
|
|
1R4B
 
 | |
1R45
 
 | | ADP-ribosyltransferase C3bot2 from Clostridium botulinum, triclinic form | | Descriptor: | GLYCEROL, Mono-ADP-ribosyltransferase C3, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L, Narumiya, S. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of ADP-ribosyltransferase C3bot2 from Clostridium botulinum
To be Published
|
|
4FML
 
 | |
4FK7
 
 | | Crystal structure of Certhrax catalytic domain | | Descriptor: | CHLORIDE ION, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Putative ADP-ribosyltransferase Certhrax, ... | | Authors: | Hong, B.S, Dimov, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-12 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
4FXQ
 
 | | Full-length Certhrax toxin from Bacillus cereus in complex with Inhibitor P6 | | Descriptor: | 8-fluoro-2-(3-piperidin-1-ylpropanoyl)-1,3,4,5-tetrahydrobenzo[c][1,6]naphthyridin-6(2H)-one, CHLORIDE ION, Putative ADP-ribosyltransferase Certhrax, ... | | Authors: | Visschedyk, D.D, Dimov, S, Kimber, M.S, Park, H.W, Merrill, A.R. | | Deposit date: | 2012-07-03 | | Release date: | 2012-09-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9599 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
4GF1
 
 | | Crystal Structure of Certhrax | | Descriptor: | Putative ADP-ribosyltransferase Certhrax, UNKNOWN ATOM OR ION | | Authors: | Hong, B.S, Dimov, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-02 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
5BWM
 
 | | The complex structure of C3cer exoenzyme and GDP bound RhoA (NADH-bound state) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADP-ribosyltransferase, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-06-08 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
6V1S
 
 | | Structure of the Clostridioides difficile transferase toxin | | Descriptor: | ADP-ribosylating binary toxin enzymatic subunit CdtA, ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Sheedlo, M.J, Anderson, D.M, Thomas, A.K, Lacy, D.B. | | Deposit date: | 2019-11-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural elucidation of theClostridioides difficiletransferase toxin reveals a single-site binding mode for the enzyme.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
