5HWN
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with pyruvate | | Descriptor: | FORMIC ACID, GLYCEROL, PYRUVIC ACID, ... | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure and function of a decarboxylating Agrobacterium tumefaciens keto-deoxy-d-galactarate dehydratase.
Biochemistry, 53, 2014
|
|
2A6N
 
 | | Dihydrodipicolinate synthase (E. coli)- mutant R138A | | Descriptor: | Dihydrodipicolinate synthase, POTASSIUM ION | | Authors: | Dobson, R.C, Devenish, S.R, Turner, L.A, Clifford, V.R, Pearce, F.G, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Role of Arginine 138 in the Catalysis and Regulation of Escherichia coli Dihydrodipicolinate Synthase.
Biochemistry, 44, 2005
|
|
2ATS
 
 | | Dihydrodipicolinate synthase co-crystallised with (S)-lysine | | Descriptor: | CHLORIDE ION, D-LYSINE, POTASSIUM ION, ... | | Authors: | Devenish, S.R.A, Dobson, R.C.J, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-08-26 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The co-crystallisation of (S)-lysine-bound dihydrodipicolinate synthase from E. coli indicates that domain movements are not responsible for (S)-lysine inhibition
To be published
|
|
6RB7
 
 | | Ruminococcus gnavus sialic acid aldolase catalytic lysine mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Bell, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-04-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut.
Nat Microbiol, 4, 2019
|
|
6RAB
 
 | |
6RD1
 
 | | Ruminococcus gnavus sialic acid aldolase catalytic lysine mutant in complex with sialic acid | | Descriptor: | 5-(acetylamino)-3,5-dideoxy-D-glycero-D-galacto-non-2-ulosonic acid, Putative N-acetylneuraminate lyase | | Authors: | Owen, C.D, Bell, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-04-12 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut.
Nat Microbiol, 4, 2019
|
|
5J5D
 
 | |
6T3T
 
 | | Structure of the 4-hydroxy-tetrahydrodipicolinate synthase from the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, SULFATE ION | | Authors: | Schmitz, R, Dietl, A, Mueller, M, Berben, T, Op den Camp, H, Barends, T. | | Deposit date: | 2019-10-11 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 4-hydroxy-tetrahydrodipicolinate synthase from the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV and the phylogeny of the aminotransferase pathway.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5KZE
 
 | | N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus | | Descriptor: | GLYCEROL, N-acetylneuraminate lyase, SULFATE ION | | Authors: | North, R.A, Watson, A.J.A, Pearce, F.G, Muscroft-Taylor, A.C, Friemann, R, Fairbanks, A.J, Dobson, R.C.J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure and inhibition of N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus.
FEBS Lett., 590, 2016
|
|
1S5T
 
 | |
1S5V
 
 | |
1S5W
 
 | |
2EHH
 
 | |
8UR1
 
 | |
6UE0
 
 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
4AHP
 
 | | Crystal Structure of Wild Type N-acetylneuraminic acid lyase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
6U01
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84D mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, L, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
8U90
 
 | |
8U92
 
 | |
8U8W
 
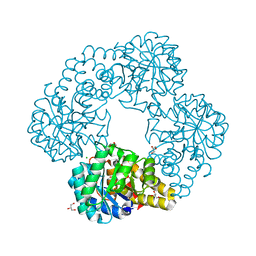 | |
8U91
 
 | |
4XKY
 
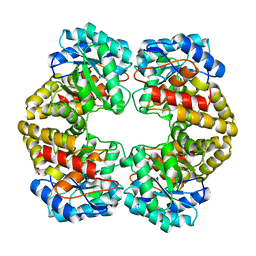 | | Structure of dihydrodipicolinate synthase from the commensal bacterium Bacteroides thetaiotaomicron at 2.1 A resolution | | Descriptor: | ACETATE ION, Dihydrodipicolinate synthase, SODIUM ION | | Authors: | Mank, N.J, Arnette, A.K, Klapper, V.G, Chruszcz, M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-01 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydrodipicolinate synthase from the commensal bacterium Bacteroides thetaiotaomicron at 2.1 angstrom resolution.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8U93
 
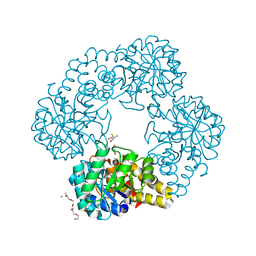 | |
6TZU
 
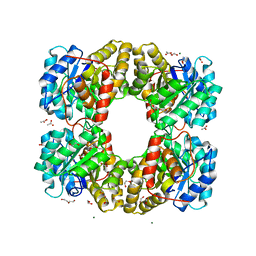 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, C, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
4UUI
 
 | |
