1Z6H
 
 | | Solution Structure of Bacillus subtilis BLAP biotinylated-form | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Biotin/Lipoyl Attachment Protein | | Authors: | Cui, G, Xia, B. | | Deposit date: | 2005-03-22 | | Release date: | 2006-03-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Identification and solution structures of a single domain biotin/lipoyl attachment protein from Bacillus subtilis
J.Biol.Chem., 281, 2006
|
|
1Z7T
 
 | |
2B8F
 
 | |
2B8G
 
 | |
2BDO
 
 | | SOLUTION STRUCTURE OF HOLO-BIOTINYL DOMAIN FROM ACETYL COENZYME A CARBOXYLASE OF ESCHERICHIA COLI DETERMINED BY TRIPLE-RESONANCE NMR SPECTROSCOPY | | Descriptor: | BIOTIN, PROTEIN (ACETYL-COA CARBOXYLASE) | | Authors: | Roberts, E.L, Shu, N, Howard, M.J, Broadhurst, R.W, Chapman-Smith, A, Wallace, J.C, Morris, T, Cronan, J.E, Perham, R.N. | | Deposit date: | 1999-03-03 | | Release date: | 1999-04-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
2D5D
 
 | |
2DNC
 
 | | Solution Structure of RSGI RUH-054, a lipoyl domain from human 2-oxoacid dehydrogenase | | Descriptor: | Pyruvate dehydrogenase protein X component | | Authors: | Ruhul Momen, A.Z.M, Hirota, H, Hayashi, F, Kurosaki, C, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-25 | | Release date: | 2006-10-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-054, a lipoyl domain from human 2-oxoacid dehydrogenase
To be published
|
|
2DN8
 
 | | Solution Structure of RSGI RUH-053, an Apo-Biotin Carboxy Carrier Protein from Human Transcarboxylase | | Descriptor: | acetyl-CoA carboxylase 2 | | Authors: | Ruhul Momen, A.Z.M, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-25 | | Release date: | 2006-10-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-053, an Apo-Biotin Carboxy Carrier Protein from Human Transcarboxylase
To be published
|
|
2DNE
 
 | | Solution Structure of RSGI RUH-058, a lipoyl domain of human 2-oxoacid dehydrogenase | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Ruhul Momen, A.Z.M, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-058, a lipoyl domain of human 2-oxoacid dehydrogenase
To be published
|
|
2EJM
 
 | | Solution structure of RUH-072, an apo-biotnyl domain form human acetyl coenzyme A carboxylase | | Descriptor: | Methylcrotonoyl-CoA carboxylase subunit alpha | | Authors: | Ruhul Momen, A.Z.M, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-19 | | Release date: | 2007-09-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RUH-072, an apo-biotnyl domain form human acetyl coenzyme A carboxylase
To be Published
|
|
2EVB
 
 | |
6ROJ
 
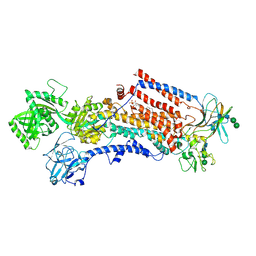 | | Cryo-EM structure of the activated Drs2p-Cdc50p | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
5CSA
 
 | |
2PNR
 
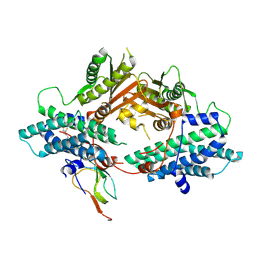 | | Crystal Structure of the asymmetric Pdk3-l2 Complex | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 3 | | Authors: | Vassylyev, D.G, Steussy, C.N, Devedjiev, Y. | | Deposit date: | 2007-04-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Asymmetric complex of Pyruvate Dehydrogenase
Kinase 3 with Lipoyl domain 2 and its Biological Implications
J.Mol.Biol., 370, 2007
|
|
2Q8I
 
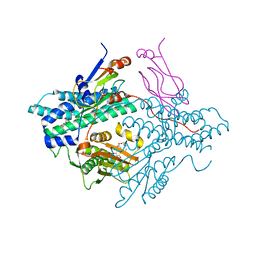 | | Pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug radicicol | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, GLYCEROL, ... | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
1Y8P
 
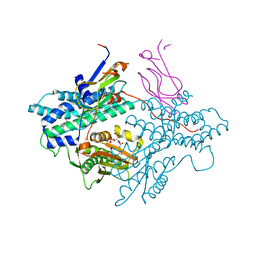 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y8N
 
 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, POTASSIUM ION, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y8O
 
 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
3BG3
 
 | |
3BG9
 
 | |
3CRL
 
 | | Crystal structure of the PDHK2-L2 complex. | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, MAGNESIUM ION, ... | | Authors: | Popov, K.M, Luo, M, Green, T.J, Grigorian, A, Klyuyeva, A, Tuganova, A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into the molecular mechanisms responsible for the regulation of pyruvate dehydrogenase kinase 2.
J.Biol.Chem., 283, 2008
|
|
3CRK
 
 | | Crystal structure of the PDHK2-L2 complex. | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, POTASSIUM ION, ... | | Authors: | Green, T.J, Popov, K.M, Luo, M, Grigorian, A, Klyuyeva, A, Tuganova, A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional insights into the molecular mechanisms responsible for the regulation of pyruvate dehydrogenase kinase 2.
J.Biol.Chem., 283, 2008
|
|
1ZY8
 
 | | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ciszak, E.M, Makal, A, Hong, Y.S, Vettaikkorumakankauv, A.K, Korotchkina, L.G, Patel, M.S. | | Deposit date: | 2005-06-09 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | How Dihydrolipoamide Dehydrogenase-binding Protein Binds Dihydrolipoamide Dehydrogenase in the Human Pyruvate Dehydrogenase Complex.
J.Biol.Chem., 281, 2006
|
|
2EJG
 
 | | Crystal Structure Of The Biotin Protein Ligase (Mutation R48A) and Biotin Carboxyl Carrier Protein Complex From Pyrococcus Horikoshii OT3 | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain, 235aa long hypothetical biotin--[acetyl-CoA-carboxylase] ligase, ADENOSINE, ... | | Authors: | Bagautdinov, B, Matsuura, Y, Bagautdinova, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
2EJF
 
 | | Crystal Structure Of The Biotin Protein Ligase (Mutations R48A and K111A) and Biotin Carboxyl Carrier Protein Complex From Pyrococcus Horikoshii OT3 | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain, 235aa long hypothetical biotin--[acetyl-CoA-carboxylase] ligase, ADENOSINE, ... | | Authors: | Bagautdinov, B, Matsuura, Y, Bagautdinova, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
