6E4B
 
 | |
6H26
 
 | | Rabbit muscle phosphoglycerate mutase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoglycerate mutase | | Authors: | Wisniewski, J, Barciszewski, J, Jaskolski, M, Rakus, D. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Rabbit muscle phosphoglycerate mutase
To Be Published
|
|
4GPI
 
 | | Crystal structure of human B type phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, Phosphoglycerate mutase 1 | | Authors: | Zhou, L, He, C. | | Deposit date: | 2012-08-21 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0817 Å) | | Cite: | Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation.
Nat Commun, 4, 2013
|
|
4GPZ
 
 | |
4HBZ
 
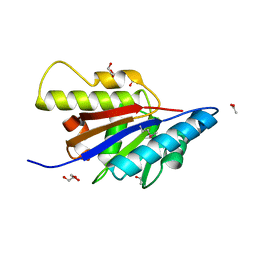 | | The Structure of Putative Phosphohistidine Phosphatase SixA from Nakamurella multipartitia. | | Descriptor: | ACETIC ACID, GLYCEROL, Putative phosphohistidine phosphatase, ... | | Authors: | Cuff, M.E, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of Putative Phosphohistidine Phosphatase SixA from Nakamurella multipartitia.
TO BE PUBLISHED
|
|
4IJ6
 
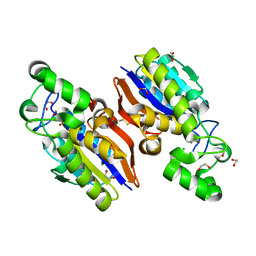 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase Mutant (H9A) from Hydrogenobacter thermophilus TK-6 in Complex with L-phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHOSERINE, ... | | Authors: | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | Deposit date: | 2012-12-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
4IJ5
 
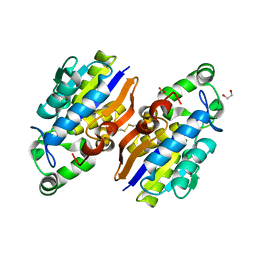 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase from Hydrogenobacter thermophilus TK-6 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoserine phosphatase 1 | | Authors: | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | Deposit date: | 2012-12-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
1BQ3
 
 | |
1BQ4
 
 | |
1C80
 
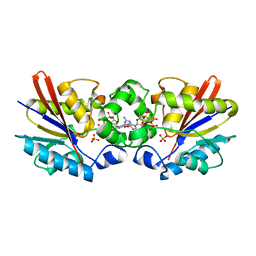 | | REGULATORY COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
1C7Z
 
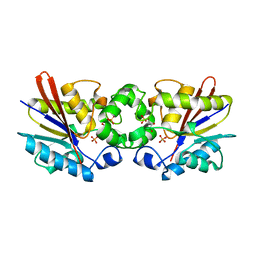 | | REGULATORY COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, GLYCERALDEHYDE-3-PHOSPHATE, PHOSPHATE ION | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
1C81
 
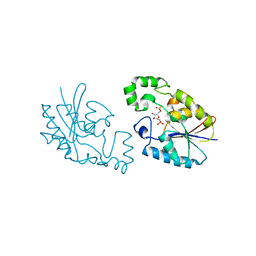 | | MICHAELIS COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1-deoxy-1-phosphono-6-O-phosphono-D-glucitol, FRUCTOSE-2,6-BISPHOSPHATASE | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
1E58
 
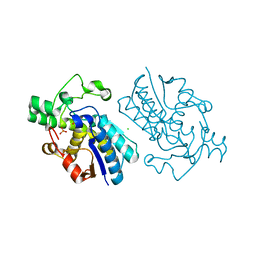 | | E.coli cofactor-dependent phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, PHOSPHOGLYCERATE MUTASE, SULFATE ION | | Authors: | Bond, C.S, Hunter, W.N. | | Deposit date: | 2000-07-19 | | Release date: | 2001-03-20 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High Resolution Structure of the Phosphohistidine-Activated Form of Escherichia Coli Cofactor-Dependent Phosphoglycerate Mutase.
J.Biol.Chem., 276, 2001
|
|
1E59
 
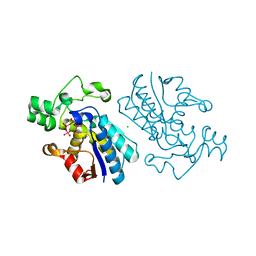 | |
1EBB
 
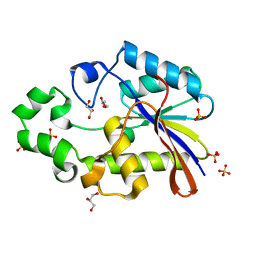 | | Bacillus stearothermophilus YhfR | | Descriptor: | GLYCEROL, PHOSPHATASE, SULFATE ION | | Authors: | Rigden, D.J, Jedrzejas, M.J. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Action of a Cofactor-Dependent Phosphoglycerate Mutase Homolog from Bacillus Stearothermophilus with Broad Specificity Phosphatase Activity.
J.Mol.Biol., 315, 2002
|
|
1FBT
 
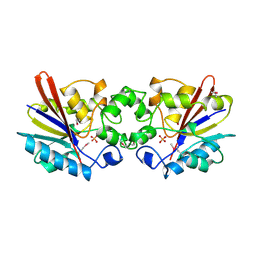 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, PHOSPHATE ION | | Authors: | Lee, Y.-H, Ogata, C, Pflugrath, J.W, Levitt, D.G, Sarma, R, Banaszak, L.J, Pilkis, S.J. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the rat liver fructose-2,6-bisphosphatase based on selenomethionine multiwavelength anomalous dispersion phases.
Biochemistry, 35, 1996
|
|
1FZT
 
 | | SOLUTION STRUCTURE AND DYNAMICS OF AN OPEN B-SHEET, GLYCOLYTIC ENZYME-MONOMERIC 23.7 KDA PHOSPHOGLYCERATE MUTASE FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | PHOSPHOGLYCERATE MUTASE | | Authors: | Uhrinova, S, Uhrin, D, Nairn, J, Price, N.C, Fothergill-Gilmore, L.A. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of an open beta-sheet, glycolytic enzyme, monomeric 23.7 kDa phosphoglycerate mutase from Schizosaccharomyces pombe.
J.Mol.Biol., 306, 2001
|
|
1H2F
 
 | |
4EO9
 
 | |
4EMB
 
 | |
3GW8
 
 | |
3GP5
 
 | |
3GP3
 
 | |
3HJG
 
 | | Crystal structure of putative alpha-ribazole-5'-phosphate phosphatase CobC FROM Vibrio parahaemolyticus | | Descriptor: | Putative alpha-ribazole-5'-phosphate phosphatase CobC, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of putative alpha-ribazole-5' -phosphate phosphatase FROM Vibrio parahaemolyticus
To be Published
|
|
3KKK
 
 | |
