3UD0
 
 | |
4F4S
 
 | |
4B2Q
 
 | |
3V3C
 
 | |
3ZO6
 
 | |
3ZK2
 
 | |
3ZK1
 
 | | Crystal structure of the sodium binding rotor ring at pH 5.3 | | Descriptor: | ATP SYNTHASE SUBUNIT C, DECYL-BETA-D-MALTOPYRANOSIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Schulz, S, Meier, T, Yildiz, O. | | Deposit date: | 2013-01-21 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A New Type of Na(+)-Driven ATP Synthase Membrane Rotor with a Two-Carboxylate Ion-Coupling Motif.
Plos Biol., 11, 2013
|
|
4MJN
 
 | |
4BEM
 
 | | Crystal structure of the F-type ATP synthase c-ring from Acetobacterium woodii. | | Descriptor: | ACETATE ION, F1FO ATPASE C1 SUBUNIT, F1FO ATPASE C2 SUBUNIT, ... | | Authors: | Matthies, D, Meier, T, Yildiz, O. | | Deposit date: | 2013-03-11 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Resolution Structure and Mechanism of an F/V-Hybrid Rotor Ring in a Na+-Coupled ATP Synthase
Nat.Commun., 5, 2014
|
|
4CBK
 
 | | The c-ring ion binding site of the ATP synthase from Bacillus pseudofirmus OF4 is adapted to alkaliphilic cell physiology | | Descriptor: | ATP SYNTHASE SUBUNIT C, DODECYL-BETA-D-MALTOSIDE, SODIUM ION, ... | | Authors: | Preiss, L, Yildiz, O, Meier, T. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The C-Ring Ion-Binding Site of the ATP Synthase from Bacillus Pseudofirmus of4 is Adapted to Alkaliphilic Lifestyle.
Mol.Microbiol., 92, 2014
|
|
4CBJ
 
 | | The c-ring ion binding site of the ATP synthase from Bacillus pseudofirmus OF4 is adapted to alkaliphilic cell physiology | | Descriptor: | ATP SYNTHASE SUBUNIT C, DODECYL-BETA-D-MALTOSIDE, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | Preiss, L, Yildiz, O, Meier, T. | | Deposit date: | 2013-10-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The C-Ring Ion-Binding Site of the ATP Synthase from Bacillus Pseudofirmus of4 is Adapted to Alkaliphilic Lifestyle.
Mol.Microbiol., 92, 2014
|
|
4UTQ
 
 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | Descriptor: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | Authors: | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | Deposit date: | 2014-07-22 | | Release date: | 2014-07-30 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
3J9T
 
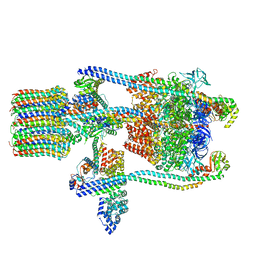 | | Yeast V-ATPase state 1 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9U
 
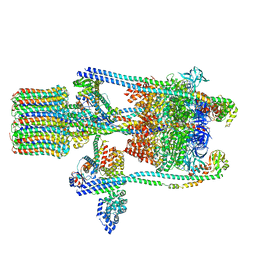 | | Yeast V-ATPase state 2 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9V
 
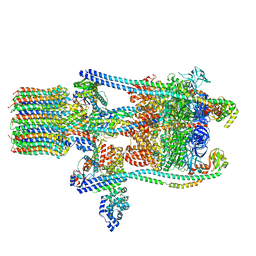 | | Yeast V-ATPase state 3 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
5BQ6
 
 | |
5BQJ
 
 | |
5BQA
 
 | |
5BPS
 
 | |
5FIL
 
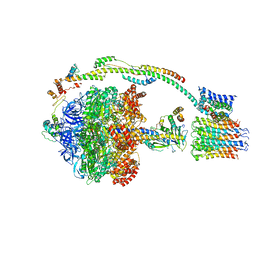 | | Bovine mitochondrial ATP synthase state 3b | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5FIJ
 
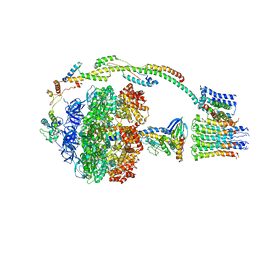 | | Bovine mitochondrial ATP synthase state 2c | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5FIK
 
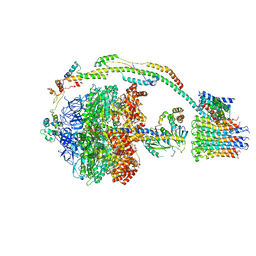 | | Bovine mitochondrial ATP synthase state 3a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5ARH
 
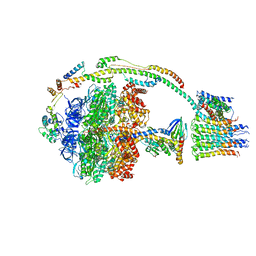 | | Bovine mitochondrial ATP synthase state 2a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5ARA
 
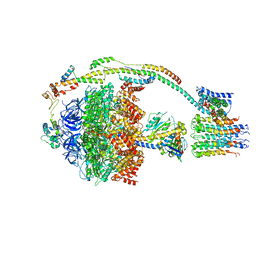 | | Bovine mitochondrial ATP synthase state 1a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5ARI
 
 | | Bovine mitochondrial ATP synthase state 2b | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
