2KMQ
 
 | |
2LEO
 
 | |
2M5X
 
 | | Novel method of protein purification for structural research. Example of ultra high resolution structure of SPI-2 inhibitor by X-ray and NMR spectroscopy. | | Descriptor: | Silk protease inhibitor 2 | | Authors: | Lenarcic Zivkovic, M, Dvornyk, A, Kludkiewicz, B, Kopera, E, Zagorski-Ostoja, W, Grzelak, K, Zhukov, I, Bal, W. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
2N71
 
 | | NMR structure of CmPI-II, a serin protease inhibitor isolated from mollusk Cenchitis muricatus | | Descriptor: | Protease inhibitor 2 | | Authors: | Cabrera-Munoz, A, Rojas, L, Alonso del Rivero Antigua, M, Pires, J. | | Deposit date: | 2015-09-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of CmPI-II, a non-classical Kazal protease inhibitor: Understanding its conformational dynamics and subtilisin A inhibition.
J.Struct.Biol., 206, 2019
|
|
2N52
 
 | |
2OVO
 
 | |
1OVO
 
 | | CRYSTALLOGRAPHIC REFINEMENT OF JAPANESE QUAIL OVOMUCOID, A KAZAL-TYPE INHIBITOR, AND MODEL BUILDING STUDIES OF COMPLEXES WITH SERINE PROTEASES | | Descriptor: | OVOMUCOID THIRD DOMAIN | | Authors: | Weber, E, Papamokos, E, Bode, W, Huber, R, Kato, I, Laskowskijunior, M. | | Deposit date: | 1982-01-18 | | Release date: | 1982-05-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic refinement of Japanese quail ovomucoid, a Kazal-type inhibitor, and model building studies of complexes with serine proteases.
J.Mol.Biol., 158, 1982
|
|
1OMU
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (REFINED MODEL USING NETWORK EDITING ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
1OMT
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (STANDARD NOESY ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
1PCE
 
 | | SOLUTION STRUCTURE AND DYNAMICS OF PEC-60, A PROTEIN OF THE KAZAL TYPE INHIBITOR FAMILY, DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | PEC-60 | | Authors: | Liepinsh, E, Berndt, K.D, Sillard, R, Mutt, V, Otting, G. | | Deposit date: | 1994-02-22 | | Release date: | 1994-04-30 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of PEC-60, a protein of the Kazal type inhibitor family, determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 239, 1994
|
|
1TUS
 
 | |
1TUR
 
 | | SOLUTION STRUCTURE OF TURKEY OVOMUCOID THIRD DOMAIN AS DETERMINED FROM NUCLEAR MAGNETIC RESONANCE DATA | | Descriptor: | OVOMUCOID | | Authors: | Krezel, A.M, Darba, P, Robertson, A.D, Fejzo, J, Macura, S, Markley, J.L. | | Deposit date: | 1994-07-06 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of turkey ovomucoid third domain as determined from nuclear magnetic resonance data.
J.Mol.Biol., 242, 1994
|
|
6KBR
 
 | |
7E50
 
 | | Crystal structure of human microplasmin in complex with kazal-type inhibitor AaTI | | Descriptor: | AAEL006007-PA, GLYCEROL, Plasminogen, ... | | Authors: | Varsha, A.W, Jobichen, C, Mok, Y.K. | | Deposit date: | 2021-02-16 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Aedes aegypti trypsin inhibitor in complex with mu-plasmin reveals role for scaffold stability in Kazal-type serine protease inhibitor.
Protein Sci., 31, 2022
|
|
3SGB
 
 | | STRUCTURE OF THE COMPLEX OF STREPTOMYCES GRISEUS PROTEASE B AND THE THIRD DOMAIN OF THE TURKEY OVOMUCOID INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | PROTEINASE B (SGPB), TURKEY OVOMUCOID INHIBITOR (OMTKY3) | | Authors: | Read, R.J, Fujinaga, M, Sielecki, A.R, James, M.N.G. | | Deposit date: | 1983-01-21 | | Release date: | 1983-07-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the complex of Streptomyces griseus protease B and the third domain of the turkey ovomucoid inhibitor at 1.8-A resolution.
Biochemistry, 22, 1983
|
|
3SGQ
 
 | | GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18,
Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
7QE9
 
 | | Human cationic trypsin (TRY1) complexed with serine protease inhibitor Kazal type 1 N34S (SPINK1 N34S) | | Descriptor: | SULFATE ION, Serine protease inhibitor Kazal-type 1, Trypsin-1 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin.
Int J Mol Sci, 23, 2022
|
|
7QE8
 
 | | Human cationic trypsin (TRY1) complexed with serine protease inhibitor Kazal type 1 (SPINK1) | | Descriptor: | SULFATE ION, Serine protease inhibitor Kazal-type 1, Trypsin-1 | | Authors: | Nagel, F, Palm, G.J, Delcea, M, Lammers, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Biophysical Insights into SPINK1 Bound to Human Cationic Trypsin.
Int J Mol Sci, 23, 2022
|
|
1AN1
 
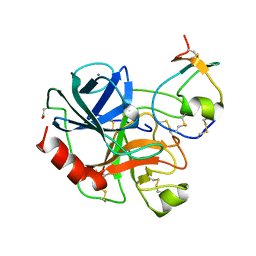 | | LEECH-DERIVED TRYPTASE INHIBITOR/TRYPSIN COMPLEX | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPTASE INHIBITOR | | Authors: | Priestle, J.P, Di Marco, S. | | Deposit date: | 1997-06-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the complex of leech-derived tryptase inhibitor (LDTI) with trypsin and modeling of the LDTI-tryptase system.
Structure, 5, 1997
|
|
1BMO
 
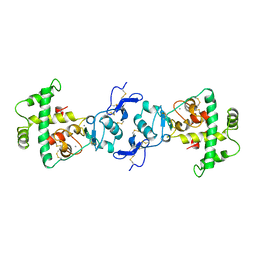 | | BM-40, FS/EC DOMAIN PAIR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BM-40, CALCIUM ION | | Authors: | Hohenester, E, Maurer, P, Timpl, R. | | Deposit date: | 1997-03-25 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a pair of follistatin-like and EF-hand calcium-binding domains in BM-40.
EMBO J., 16, 1997
|
|
2F3C
 
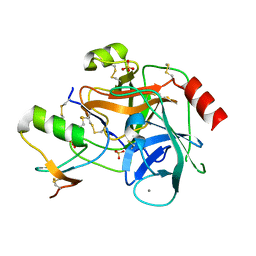 | | Crystal structure of infestin 1, a Kazal-type serineprotease inhibitor, in complex with trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Campos, I.T.N, Tanaka, A.S, Barbosa, J.A.R.G. | | Deposit date: | 2005-11-20 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Kazal-type inhibitors infestins 1 and 4 differ in specificity but are similar in three-dimensional structure.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1DS2
 
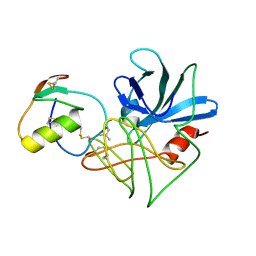 | | CRYSTAL STRUCTURE OF SGPB:OMTKY3-COO-LEU18I | | Descriptor: | OVOMUCOID, PROTEINASE B (SGPB) | | Authors: | Bateman, K.S, Huang, K, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2000-01-06 | | Release date: | 2001-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
1HJA
 
 | |
1YU6
 
 | | Crystal Structure of the Subtilisin Carlsberg:OMTKY3 Complex | | Descriptor: | CALCIUM ION, Ovomucoid, Subtilisin Carlsberg | | Authors: | Maynes, J.T, Cherney, M.M, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2005-02-11 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the subtilisin Carlsberg-OMTKY3 complex reveals two different ovomucoid conformations.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1CT4
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-VAL18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
