7TBB
 
 | | Crystal structure of Plasmepsin X from Plasmodium falciparum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Christensen, J.B, Hodder, A.N, Dietrich, M.H, Scally, S.W, Cowman, A.F. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Basis for drug selectivity of plasmepsin IX and X inhibition in Plasmodium falciparum and vivax.
Structure, 30, 2022
|
|
7TBE
 
 | | Crystal structure of Plasmepsin X from Plasmodium vivax in complex with WM4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(R)-[(2E,4S)-2-imino-4-methyl-6-oxo-4-(propan-2-yl)-1,3-diazinan-1-yl](phenyl)methyl]-N-[(1S)-1-phenylethyl]benzamide, Plasmepsin X, ... | | Authors: | Hodder, A.N, Christensen, J.B, Scally, S.W, Cowman, A.F. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Basis for drug selectivity of plasmepsin IX and X inhibition in Plasmodium falciparum and vivax.
Structure, 30, 2022
|
|
7TBC
 
 | | Crystal structure of Plasmepsin X from Plasmodium falciparum in complex with WM382 | | Descriptor: | (4R)-4-[(2E)-4,4-diethyl-2-imino-6-oxo-1,3-diazinan-1-yl]-N-[(4S)-2,2-dimethyl-3,4-dihydro-2H-1-benzopyran-4-yl]-3,4-dihydro-2H-1-benzopyran-6-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Christensen, J.B, Hodder, A.N, Scally, S.W, Cowman, A.F. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Basis for drug selectivity of plasmepsin IX and X inhibition in Plasmodium falciparum and vivax.
Structure, 30, 2022
|
|
6OD6
 
 | | Structure of BACE-1 in complex with Ligand 13 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{3-[(3R)-1-amino-3-methyl-3,4-dihydropyrrolo[1,2-a]pyrazin-3-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Authors: | Shaffer, P.L. | | Deposit date: | 2019-03-26 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluation of a Series of beta-Secretase 1 Inhibitors Containing Novel Heteroaryl-Fused-Piperazine Amidine Warheads.
Acs Med.Chem.Lett., 10, 2019
|
|
7NKW
 
 | | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-02-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector
To Be Published
|
|
6PZ4
 
 | | co-crystal structure of BACE with inhibitor AM-6494 | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Huang, X. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-23 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of AM-6494: A Potent and Orally Efficacious beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitor with in Vivo Selectivity over BACE2.
J.Med.Chem., 63, 2020
|
|
6QBG
 
 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 14 | | Descriptor: | (3~{S},7~{S},8~{S})-8-(naphthalen-2-ylmethyl)-7-oxidanyl-3-propan-2-yl-1,4,9-triazacyclohenicosane-2,5,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
7VE0
 
 | | Crystal Structure of Ritonavir bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2021-09-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of Plasmodium falciparum plasmepsins by drugs targeting HIV-1 protease: A way forward for antimalarial drug discovery.
Curr Res Struct Biol, 7, 2024
|
|
7VE2
 
 | | Crystal Structure of Lopinavir bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, Plasmepsin II | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2021-09-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Plasmodium falciparum plasmepsins by drugs targeting HIV-1 protease: A way forward for antimalarial drug discovery.
Curr Res Struct Biol, 7, 2024
|
|
6ZZ2
 
 | | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Hassaan, E, Klebe, G, Heine, A, Schiebel, J, Koester, H. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14885437 Å) | | Cite: | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin
To Be Published
|
|
7ADG
 
 | | Cocktail experiment G: fragments 216 and 338 at 90 mM concentration in complex with Endothiapepsin | | Descriptor: | 2-(1H-imidazol-1-yl)-N-(trans-4-methylcyclohexyl)acetamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Hassaan, E, Klebe, G, Heine, A, Schiebel, J, Koester, H. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.0794636 Å) | | Cite: | Comparison of Cocktail Screening in X-Ray Crystallography vs NMR
To Be Published
|
|
6DHC
 
 | | X-ray structure of BACE1 in complex with a bicyclic isoxazoline carboxamide as the P3 ligand | | Descriptor: | (3R,3aR,6aS)-N-[(4R,7S,8S,10R,13S)-8-hydroxy-10,17-dimethyl-7-(2-methylpropyl)-5,11,14-trioxo-13-(propan-2-yl)-2-thia-6,12,15-triazaoctadecan-4-yl]hexahydrofuro[3,2-d][1,2]oxazole-3-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Mesecar, A.D, Lendy, E.K. | | Deposit date: | 2018-05-19 | | Release date: | 2018-07-25 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design, synthesis, X-ray studies, and biological evaluation of novel BACE1 inhibitors with bicyclic isoxazoline carboxamides as the P3 ligand.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6DMI
 
 | | A multiconformer ligand model of 5T5 bound to BACE-1 | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6E3Z
 
 | | Structure of Bace-1 in complex with Ligand 8 | | Descriptor: | Beta-secretase 1, N-{3-[(2R,3R)-5-amino-3-methyl-2-(trifluoromethyl)-3,6-dihydro-2H-1,4-oxazin-3-yl]-4-fluorophenyl}-3,5-dichloropyridine-2-carboxamide | | Authors: | Shaffer, P.L. | | Deposit date: | 2018-07-16 | | Release date: | 2019-09-11 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and Chemical Development of JNJ-50138803, a Clinical Candidate BACE1 Inhibitor
Acs Symp.Ser., 1307, 2020
|
|
6EJ2
 
 | | BACE1 compound 28 | | Descriptor: | Beta-secretase 1, compound 28 | | Authors: | Johansson, P. | | Deposit date: | 2017-09-20 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Toward beta-Secretase-1 Inhibitors with Improved Isoform Selectivity.
J. Med. Chem., 61, 2018
|
|
6EJ3
 
 | | BACE1 compound 23 | | Descriptor: | (1r,4r)-4-methoxy-6'-(5-methyl-3-pyridinyl)-3'H-dispiro[cyclohexane-1,2'-indene-1',4''-[1,3]oxazol]-2''-amine, Beta-secretase 1 | | Authors: | Johansson, P. | | Deposit date: | 2017-09-20 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Toward beta-Secretase-1 Inhibitors with Improved Isoform Selectivity.
J. Med. Chem., 61, 2018
|
|
6EQM
 
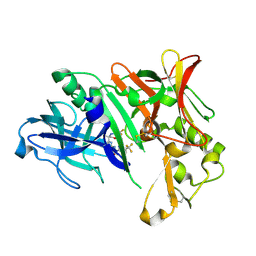 | | Crystal Structure of Human BACE-1 in Complex with CNP520 | | Descriptor: | Beta-secretase 1, ~{N}-[6-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]-5-fluoranyl-pyridin-2-yl]-3-chloranyl-5-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Rondeau, J.-M, Wirth, E. | | Deposit date: | 2017-10-13 | | Release date: | 2018-09-19 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The BACE-1 inhibitor CNP520 for prevention trials in Alzheimer's disease.
EMBO Mol Med, 10, 2018
|
|
6FGY
 
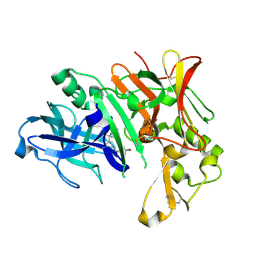 | | Crystal Structure of Human BACE-1 in Complex with amino-1,4-oxazine compound 4 | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R})-5-azanyl-3-methyl-2,6-dihydro-1,4-oxazin-3-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.-M, Bourgier, E. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-06 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of amino-1,4-oxazines as potent BACE-1 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
1BBS
 
 | |
1APV
 
 | |
1BIM
 
 | | CRYSTALLOGRAPHIC STUDIES ON THE BINDING MODES OF P2-P3 BUTANEDIAMIDE RENIN INHIBITORS | | Descriptor: | (2S)-2-[(2-amino-1,3-thiazol-4-yl)methyl]-N~1~-{(1S,2S)-1-(cyclohexylmethyl)-2-hydroxy-2-[(3R)-1,5,5-trimethyl-2-oxopyrrolidin-3-yl]ethyl}-N~4~-[2-(dimethylamino)-2-oxoethyl]-N~4~-[(1S)-1-phenylethyl]butanediamide, Renin | | Authors: | Tong, L. | | Deposit date: | 1995-09-27 | | Release date: | 1996-01-29 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on the binding modes of P2-P3 butanediamide renin inhibitors.
J.Biol.Chem., 270, 1995
|
|
6UJ0
 
 | | Unbound BACE2 mutant structure | | Descriptor: | Beta-secretase 2, unidentified polypeptide | | Authors: | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Structure-Based Discovery Platform for BACE2 and the Development of Selective BACE Inhibitors.
Acs Chem Neurosci, 12, 2021
|
|
1B5F
 
 | | NATIVE CARDOSIN A FROM CYNARA CARDUNCULUS L. | | Descriptor: | PROTEIN (CARDOSIN A), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Frazao, C, Bento, I, Carrondo, M.A. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of cardosin A, a glycosylated and Arg-Gly-Asp-containing aspartic proteinase from the flowers of Cynara cardunculus L.
J.Biol.Chem., 274, 1999
|
|
1BIL
 
 | | CRYSTALLOGRAPHIC STUDIES ON THE BINDING MODES OF P2-P3 BUTANEDIAMIDE RENIN INHIBITORS | | Descriptor: | (2S)-2-[(2-amino-1,3-thiazol-4-yl)methyl]-N~1~-[(1S,2R,3R)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methylhexyl]-N~4~-[2-(d imethylamino)-2-oxoethyl]-N~4~-[(1S)-1-phenylethyl]butanediamide, Renin | | Authors: | Tong, L. | | Deposit date: | 1995-09-27 | | Release date: | 1996-01-29 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies on the binding modes of P2-P3 butanediamide renin inhibitors.
J.Biol.Chem., 270, 1995
|
|
6UJ1
 
 | | BACE2 mutant in complex with a macrocyclic compound | | Descriptor: | (3S)-3-hydroxy-N-(2-methylpropyl)-N~2~-{[(4S)-17-[(methylsulfonyl)(propyl)amino]-2-oxo-3-azatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaen-4-yl]methyl}-L-norleucinamide, Beta-secretase 2 | | Authors: | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | Deposit date: | 2019-10-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | A Structure-Based Discovery Platform for BACE2 and the Development of Selective BACE Inhibitors.
Acs Chem Neurosci, 12, 2021
|
|
