2VAV
 
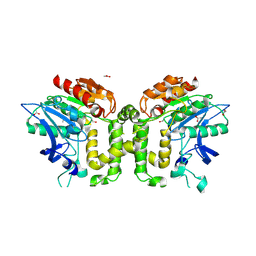 | | Crystal structure of deacetylcephalosporin C acetyltransferase (DAC-Soak) | | Descriptor: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The last step in cephalosporin C formation revealed: crystal structures of deacetylcephalosporin C acetyltransferase from Acremonium chrysogenum in complexes with reaction intermediates.
J. Mol. Biol., 377, 2008
|
|
2VAT
 
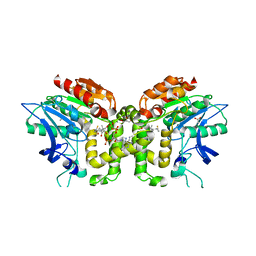 | | Crystal structure of deacetylcephalosporin C acetyltransferase in complex with coenzyme A | | Descriptor: | ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE, COENZYME A, ... | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Last Step in Cephalosporin C Formation Revealed: Crystal Structures of Deacetylcephalosporin C Acetyltransferase from Acremonium Chrysogenum in Complexes with Reaction Intermediates.
J.Mol.Biol., 377, 2008
|
|
7WAM
 
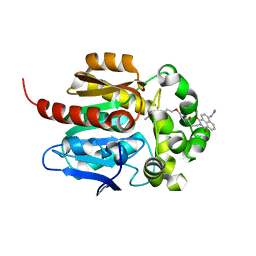 | | Crystal structure of HaloTag complexed with VL1 | | Descriptor: | 3-[6-(2-azanylhydrazinyl)-1,3-bis(oxidanylidene)benzo[de]isoquinolin-2-yl]-N-[2-(2-hexoxyethoxy)ethyl]propanamide, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Pratyush, M, Kang, M, Lee, H, Lee, C, Rhee, H. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A chemical tool for blue light-inducible proximity photo-crosslinking in live cells.
Chem Sci, 13, 2022
|
|
7WAN
 
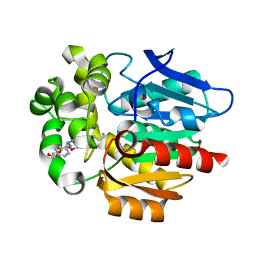 | | Crystal structure of HaloTag complexed with UL2 | | Descriptor: | (R)-[4-(2-azanylhydrazinyl)phenyl]-[2-[2-(2-hexoxyethoxy)ethoxy]ethylamino]methanol, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Pratyush, M, Kang, M, Lee, H, Lee, C, Rhee, H. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | A chemical tool for blue light-inducible proximity photo-crosslinking in live cells.
Chem Sci, 13, 2022
|
|
2V9Z
 
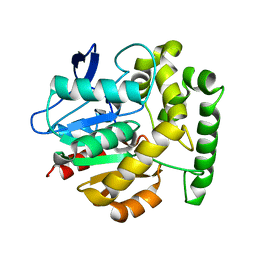 | | Structure of the Rhodococcus haloalkane dehalogenase mutant with enhanced enantioselectivity | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Koudelakova, T, Prokop, Z, Sato, Y, Lapkouski, M, Chovancova, E, Monincova, M, Jesenska, A, Emmer, J, Senda, T, Nagata, Y, Kuta Smatanova, I, Damborsky, J. | | Deposit date: | 2007-08-28 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rational Engineering of Rhodococcus Haloalkane Dehalogenase with Enhanced Enantioselectivity
To be Published
|
|
2VF2
 
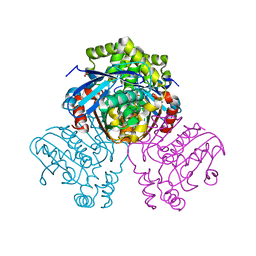 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2WFM
 
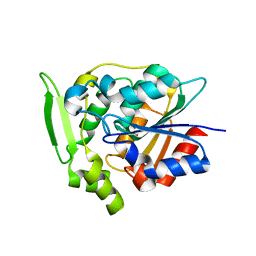 | | Crystal structure of polyneuridine aldehyde esterase mutant (H244A) | | Descriptor: | POLYNEURIDINE ALDEHYDE ESTERASE | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2WFL
 
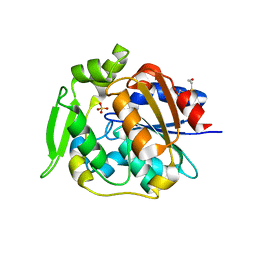 | | Crystal structure of polyneuridine aldehyde esterase | | Descriptor: | POLYNEURIDINE-ALDEHYDE ESTERASE, SULFATE ION | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2WJ3
 
 | |
2WJ6
 
 | |
2WJ4
 
 | |
2WM2
 
 | |
2WUF
 
 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with 4,9DSHA | | Descriptor: | (3E,5R)-8-[(1S,3AR,4R,7AS)-1-HYDROXY-7A-METHYL-5-OXOOCTAHYDRO-1H-INDEN-4-YL]-5-METHYL-2,6-DIOXOOCT-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, ... | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism.
J. Biol. Chem., 285, 2010
|
|
7YAS
 
 | | HYDROXYNITRILE LYASE, LOW TEMPERATURE NATIVE STRUCTURE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, PROTEIN (HYDROXYNITRILE LYASE), ... | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
2WUD
 
 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, THIOCYANATE ION | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism.
J. Biol. Chem., 285, 2010
|
|
2WUE
 
 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with HOPODA | | Descriptor: | (3E,5R)-8-(2-CHLOROPHENYL)-5-METHYL-2,6-DIOXOOCT-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, THIOCYANATE ION | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism.
J. Biol. Chem., 285, 2010
|
|
2WUG
 
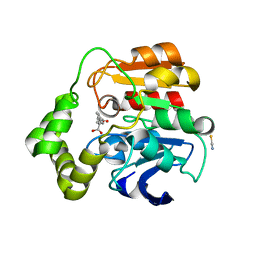 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, ... | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R.L, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a C-C Hydrolase from Mycobacterium Tuberuclosis Involved in Cholesterol Metabolism.
J.Biol.Chem., 285, 2010
|
|
2XUA
 
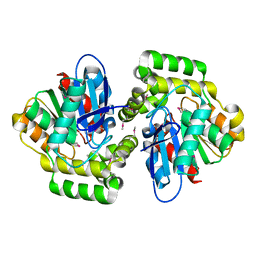 | | Crystal structure of the enol-lactonase from Burkholderia xenovorans LB400 | | Descriptor: | 3-OXOADIPATE ENOL-LACTONASE, LAEVULINIC ACID | | Authors: | Bains, J, Kaufman, L, Farnell, B, Boulanger, M.J. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Product Analog Bound Form of 3-Oxoadipate-Enol- Lactonase (Pcad) Reveals a Multifunctional Role for the Divergent CAP Domain.
J.Mol.Biol., 406, 2011
|
|
2XMZ
 
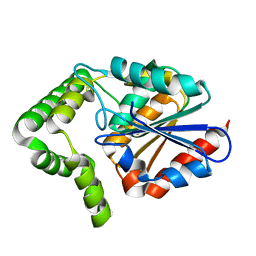 | | Structure of MenH from S. aureus | | Descriptor: | HYDROLASE, ALPHA/BETA HYDROLASE FOLD FAMILY | | Authors: | Dawson, A, Fyfe, P.K, Gillet, F, Hunter, W.N. | | Deposit date: | 2010-07-30 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Exploiting the High-Resolution Crystal Structure of Staphylococcus Aureus Menh to Gain Insight Into Enzyme Activity.
Bmc Struct.Biol., 11, 2011
|
|
2XT0
 
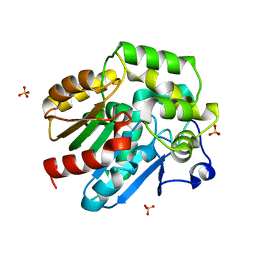 | | Dehalogenase DPpA from Plesiocystis pacifica SIR-I | | Descriptor: | HALOALKANE DEHALOGENASE, SULFATE ION | | Authors: | Bogdanovic, X, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-10-02 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cloning, Functional Expression, Biochemical Characterization, and Structural Analysis of a Haloalkane Dehalogenase from Plesiocystis Pacifica Sir-1.
Appl.Microbiol.Biotechnol., 91, 2011
|
|
7ZM1
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclophostin-like inhibitor CyC7b | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, methoxy-[(~{E},3~{R})-3-[(2~{R})-1-methoxy-1,3-bis(oxidanylidene)butan-2-yl]tridec-11-enyl]phosphinous acid | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7ZM3
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclipostin-like inhibitor CyC17 | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, hexadecyl dihydrogen phosphate | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7ZJT
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis at 1.96 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, CHLORIDE ION | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7ZM2
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclophostin-like inhibitor CyC8b | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, methoxy-[(3~{R})-3-[(2~{R})-1-methoxy-1,3-bis(oxidanylidene)butan-2-yl]pentadecyl]phosphinic acid | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7ZM4
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclipostin-like inhibitor CyC31 | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, undecyl dihydrogen phosphate | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
