1WBE
 
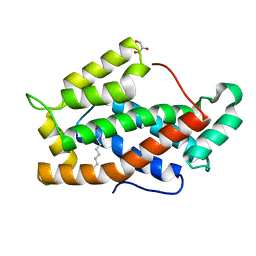 | | X-ray structure of bovine GLTP | | Descriptor: | DECANOIC ACID, GLYCEROL, GLYCOLIPID TRANSFER PROTEIN | | Authors: | Airenne, T.T, Kidron, H, West, G, Nymalm, Y, Nylund, M, Mattjus, P, Salminen, T.A. | | Deposit date: | 2004-11-01 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Evidence for Adaptive Ligand Binding of Glycolipid Transfer Protein.
J.Mol.Biol., 355, 2006
|
|
5KDI
 
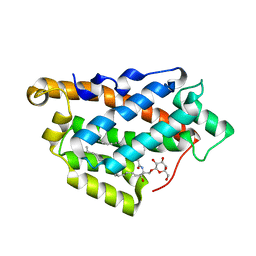 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | Descriptor: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | Authors: | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2016-06-08 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
3KV0
 
 | | Crystal structure of HET-C2: A FUNGAL GLYCOLIPID TRANSFER PROTEIN (GLTP) | | Descriptor: | HET-C2 | | Authors: | Simanshu, D.K, Kenoth, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2009-11-28 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determination and tryptophan fluorescence of heterokaryon incompatibility C2 protein (HET-C2), a fungal glycolipid transfer protein (GLTP), provide novel insights into glycolipid specificity and membrane interaction by the GLTP fold.
J.Biol.Chem., 285, 2010
|
|
1SWX
 
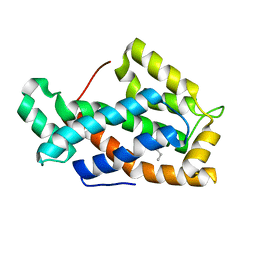 | | Crystal structure of a human glycolipid transfer protein in apo-form | | Descriptor: | Glycolipid transfer protein, HEXANE | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for glycosphingolipid transfer specificity.
Nature, 430, 2004
|
|
1SX6
 
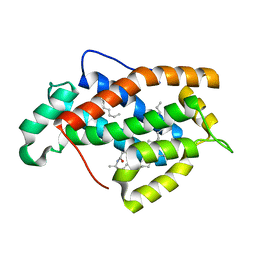 | | Crystal structure of human Glycolipid Transfer protein in lactosylceramide-bound form | | Descriptor: | Glycolipid transfer protein, N-OCTANE, OLEIC ACID, ... | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for glycosphingolipid transfer specificity.
Nature, 430, 2004
|
|
1TFJ
 
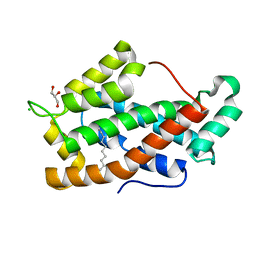 | | Crystal structure of Bovine Glycolipid transfer protein in complex with a fatty acid | | Descriptor: | CHLORIDE ION, DECANOIC ACID, GLYCEROL, ... | | Authors: | Airenne, T.T, Kidron, H, West, G, Nymalm, Y, Mattjus, P, Salminen, T.A. | | Deposit date: | 2004-05-27 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural evidence for adaptive ligand binding of glycolipid transfer protein.
J.Mol.Biol., 355, 2006
|
|
4NTO
 
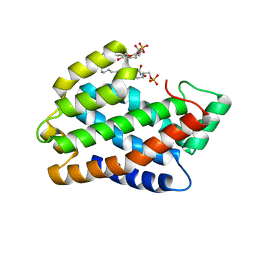 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C2 ceramide-1-phosphate (d18:1/2:0) at 2.15 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NT1
 
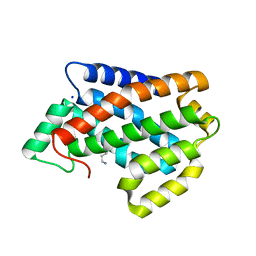 | | Crystal structure of apo-form of Arabidopsis ACD11 (accelerated-cell-death 11) at 1.8 Angstrom resolution | | Descriptor: | SODIUM ION, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NT2
 
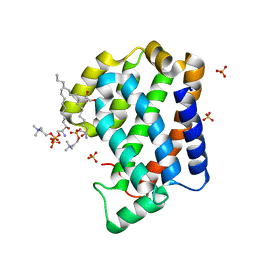 | | Crystal structure of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with lyso-sphingomyelin (d18:1) at 2.4 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(R)-{[(2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl]oxy}(hydroxy)phosphoryl]oxy}-N,N,N-trimethylethanaminium, SULFATE ION, ... | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NTI
 
 | | Crystal structure of D60N mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.9 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NTG
 
 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.55 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5505 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
3RIC
 
 | | Crystal Structure of D48V||A47D mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3RWV
 
 | | Crystal Structure of apo-form of Human Glycolipid Transfer Protein at 1.5 A resolution | | Descriptor: | Glycolipid transfer protein, SULFATE ION | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3S0K
 
 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with glucosylceramide containing oleoyl acyl chain (18:1) | | Descriptor: | (9Z)-N-[(2S,3R,4E)-1-(beta-D-glucopyranosyloxy)-3-hydroxyoctadec-4-en-2-yl]octadec-9-enamide, Glycolipid transfer protein, NICKEL (II) ION | | Authors: | Cabo-Bilbao, A, Samygina, V, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3S0I
 
 | | Crystal Structure of D48V mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo galactosylceramide containing nervonoyl acyl chain | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Popov, A.N, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3RZN
 
 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
2BV7
 
 | | Crystal structure of GLTP with bound GM3 | | Descriptor: | GLYCOLIPID TRANSFER PROTEIN, N-{1-[(HEXOPYRANOSYLOXY)METHYL]-2-HYDROXYNONADECYL}TETRACOSANAMIDE, SULFATE ION | | Authors: | Kidron, H, Airenne, T.T, Nymalm, Y, Nylund, M, West, G, Mattjus, P, Salminen, T.A. | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Evidence for Adaptive Ligand Binding of Glycolipid Transfer Protein.
J.Mol.Biol., 355, 2006
|
|
2EVD
 
 | | Crystal structure of human Glycolipid Transfer Protein complexed with 12:0 Lactosylceramide | | Descriptor: | DECANE, Glycolipid transfer protein, LAURIC ACID, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EVL
 
 | | Crystal structure of human Glycolipid Transfer Protein complexed with 18:2 Galactosylceramide | | Descriptor: | Glycolipid transfer protein, LINOLEIC ACID, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EUM
 
 | | Crystal structure of human Glycolipid Transfer Protein complexed with 8:0 Lactosylceramide | | Descriptor: | DECANE, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EVS
 
 | | Crystal structure of human Glycolipid Transfer Protein complexed with n-hexyl-beta-D-glucoside | | Descriptor: | DECANE, Glycolipid transfer protein, HEXANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EUK
 
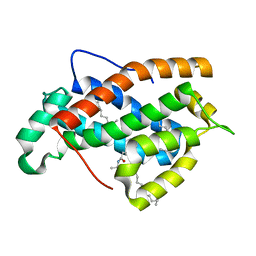 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 24:1 Galactosylceramide | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EVT
 
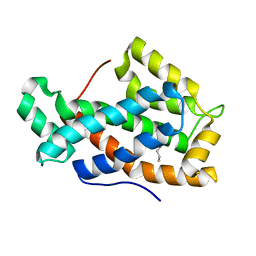 | | Crystal structure of D48V mutant of human Glycolipid Transfer Protein | | Descriptor: | Glycolipid transfer protein, HEXANE | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
4GIX
 
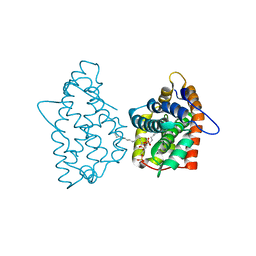 | | Crystal structure of human GLTP bound with 12:0 disulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | Deposit date: | 2012-08-09 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHS
 
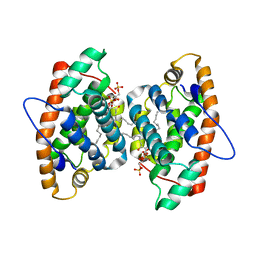 | | Crystal structure of human GLTP bound with 12:0 disulfatide (orthorombic form; two subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Popov, A.N, Goni-de-Cerio, F, Cabo-Bilbao, A, Malinina, L. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
