8DQV
 
 | | The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.52 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
8W6X
 
 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
5D51
 
 | | Krypton derivatization of an O2-tolerant membrane-bound [NiFe] hydrogenase reveals a hydrophobic gas tunnel network | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Kalms, J, Schmidt, A, Frielingsdorf, S, van der Linden, P, von Stetten, D, Lenz, O, Carpentier, P, Scheerer, P. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Krypton Derivatization of an O2 -Tolerant Membrane-Bound [NiFe] Hydrogenase Reveals a Hydrophobic Tunnel Network for Gas Transport.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1CC1
 
 | | CRYSTAL STRUCTURE OF A REDUCED, ACTIVE FORM OF THE NI-FE-SE HYDROGENASE FROM DESULFOMICROBIUM BACULATUM | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROGENASE (LARGE SUBUNIT), ... | | Authors: | Garcin, E, Vernede, X, Hatchikian, E.C, Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1999-03-03 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a reduced [NiFeSe] hydrogenase provides an image of the activated catalytic center
Structure Fold.Des., 7, 1999
|
|
4OMF
 
 | | The F420-reducing [NiFe]-hydrogenase complex from Methanothermobacter marburgensis, the first X-ray structure of a group 3 family member | | Descriptor: | CHLORIDE ION, F420-reducing hydrogenase, subunit alpha, ... | | Authors: | Vitt, S, Ma, K, Warkentin, E, Moll, J, Pierik, A, Shima, S, Ermler, U. | | Deposit date: | 2014-01-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The F420-Reducing [NiFe]-Hydrogenase Complex from Methanothermobacter marburgensis, the First X-ray Structure of a Group 3 Family Member.
J.Mol.Biol., 426, 2014
|
|
1UBM
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
2FRV
 
 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF NI-FE HYDROGENASE | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1997-06-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of the [Nife] Hydrogenase Active Site: Evidence for Biologically Uncommon Fe Ligands
J.Am.Chem.Soc., 118, 1996
|
|
3MYR
 
 | | Crystal structure of [NiFe] hydrogenase from Allochromatium vinosum in its Ni-A state | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Hydrogenase (NiFe) small subunit HydA, ... | | Authors: | Ogata, H, Kellers, P, Lubitz, W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the [NiFe] hydrogenase from the photosynthetic bacterium Allochromatium vinosum: characterization of the oxidized enzyme (Ni-A state).
J.Mol.Biol., 402, 2010
|
|
2WPN
 
 | | Structure of the oxidised, as-isolated NiFeSe hydrogenase from D. vulgaris Hildenborough | | Descriptor: | 3-[DODECYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Marques, M.C, Coelho, R, De Lacey, A.L, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-01-12 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The three-dimensional structure of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough: a hydrogenase without a bridging ligand in the active site in its oxidised, "as-isolated" state.
J.Mol.Biol., 396, 2010
|
|
4C3O
 
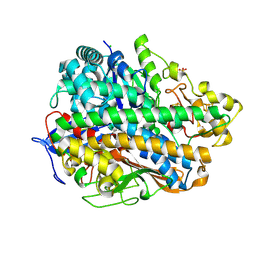 | | Structure and function of an oxygen tolerant NiFe hydrogenase from Salmonella | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Bowman, L, Flanagan, L, Fyfe, P.K, Parkin, A, Hunter, W.N, Sargent, F. | | Deposit date: | 2013-08-26 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How the Structure of the Large Subunit Controls Function in an Oxygen-Tolerant [Nife]-Hydrogenase.
Biochem.J., 458, 2014
|
|
4CI0
 
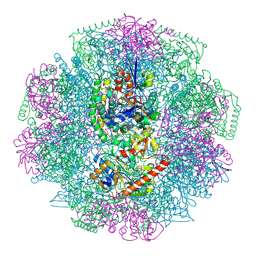 | | Electron cryo-microscopy of F420-reducing NiFe hydrogenase Frh | | Descriptor: | F420-REDUCING HYDROGENASE, SUBUNIT ALPHA, SUBUNIT BETA, ... | | Authors: | Allegretti, M, Mills, D.J, McMullan, G, Kuehlbrandt, W, Vonck, J. | | Deposit date: | 2013-12-05 | | Release date: | 2014-02-26 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Atomic Model of the F420-Reducing [Nife] Hydrogenase by Electron Cryo-Electron Microscopy Using a Direct Electron Detector.
Elife, 3, 2014
|
|
3RGW
 
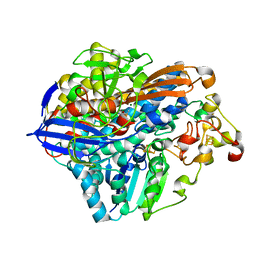 | | Crystal structure at 1.5 A resolution of an H2-reduced, O2-tolerant hydrogenase from Ralstonia eutropha unmasks a novel iron-sulfur cluster | | Descriptor: | FE3-S4 CLUSTER, FE4-S3 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Scheerer, P, Fritsch, J, Frielingsdorf, S, Kroschinsky, S, Friedrich, B, Lenz, O, Spahn, C.M.T. | | Deposit date: | 2011-04-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre.
Nature, 479, 2011
|
|
3AYX
 
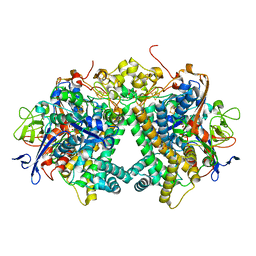 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an H2-reduced condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
4UQL
 
 | | High-resolution structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystallographic Studies of [Nife]-Hydrogenase Mutants: Towards Consensus Structures for the Elusive Unready Oxidized States.
J.Biol.Inorg.Chem., 20, 2015
|
|
4UQP
 
 | | High-resolution structure of the D. fructosovorans NiFe-hydrogenase L122A mutant after exposure to air | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic Studies of [Nife]-Hydrogenase Mutants: Towards Consensus Structures for the Elusive Unready Oxidized States.
J.Biol.Inorg.Chem., 20, 2015
|
|
4URH
 
 | | High-resolution structure of partially oxidized D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
5YY0
 
 | | Crystal structure of the HyhL-HypA complex (form II) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.243 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YXY
 
 | | Crystal structure of the HyhL-HypA complex (form I) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7NEM
 
 | | Hydrogenase-2 variant R479K - anaerobically oxidised form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B. | | Deposit date: | 2021-02-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A comprehensive structural and kinetic investigation of the role of the active-site argininein bidirectional hydrogen activation by the [NiFe]-hydrogenase "Hyd-2) from Escherichia coli
To Be Published
|
|
7ODH
 
 | |
7ODG
 
 | |
8POV
 
 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the H2-reduced state at 1.92 A Resolution. | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POU
 
 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.65 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POW
 
 | | Crystal Structure of the C19G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.61 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Fe4S4, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
3AYZ
 
 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an air-oxidized condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
