6M10
 
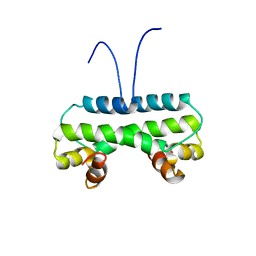 | | Crystal structure of PA4853 (Fis) from Pseudomonas aeruginosa | | Descriptor: | Putative Fis-like DNA-binding protein | | Authors: | Zhang, H, Gao, Z, Zhou, J, Dong, Y. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal structure of the nucleoid-associated protein Fis (PA4853) from Pseudomonas aeruginosa.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4FIS
 
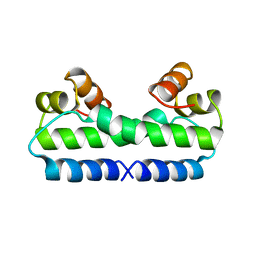 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J.-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1NTC
 
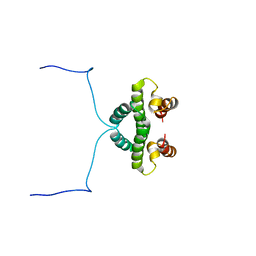 | |
2M8G
 
 | |
3E7L
 
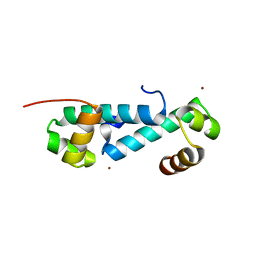 | | Crystal structure of sigma54 activator NtrC4's DNA binding domain | | Descriptor: | Transcriptional regulator (NtrC family), ZINC ION | | Authors: | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
3FIS
 
 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
4FTH
 
 | | Crystal Structure of NtrC4 DNA-binding domain bound to double-stranded DNA | | Descriptor: | 5'-D(*AP*CP*TP*TP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*AP*AP*AP*TP*GP*CP*AP*T)-3', 5'-D(P*GP*AP*TP*GP*CP*AP*TP*TP*TP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*AP*A)-3', Transcriptional regulator (NtrC family) | | Authors: | Vidangos, N.K, Heideker, J, Lyubimov, A.Y, Lamers, M, Huo, Y, Pelton, J.G, Ton, J, Gralla, J.D, Kuriyan, J, Berger, J.M, Wemmer, D.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | DNA Recognition by a sigma (54) Transcriptional Activator from Aquifex aeolicus.
J.Mol.Biol., 426, 2014
|
|
4L5E
 
 | | Crystal structure of A. aeolicus NtrC1 DNA binding domain | | Descriptor: | SULFATE ION, Transcriptional regulator (NtrC family) | | Authors: | Young, A, Maris, A.E, Vidangos, N.K, Hong, E, Pelton, J.G, Batchelor, J.D, Wemmer, D.E. | | Deposit date: | 2013-06-10 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure, function, and tethering of DNA-binding domains in sigma (54) transcriptional activators.
Biopolymers, 99, 2013
|
|
5DTD
 
 | |
5E3L
 
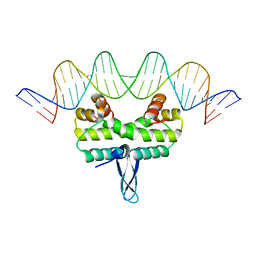 | |
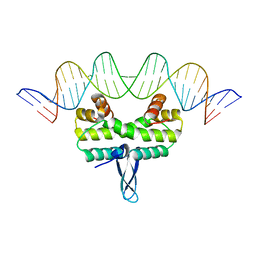
5DS9
 
 | |
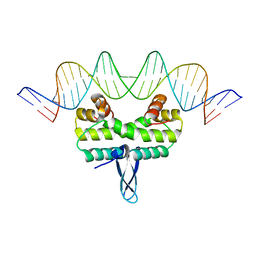
5E3O
 
 | |
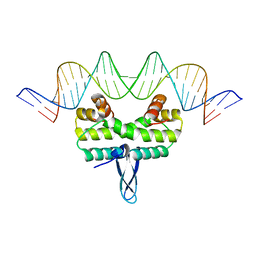
5E3N
 
 | |
5E3M
 
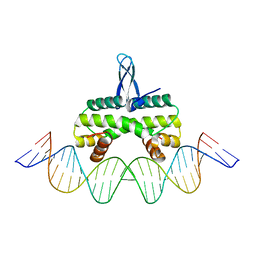 | | Crystal structure of Fis bound to 27bp DNA F35 (AAATTAGTTTGAATCTCGAGCTAATTT) | | Descriptor: | DNA (27-MER), DNA-binding protein Fis | | Authors: | Stella, S, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2015-10-03 | | Release date: | 2016-03-09 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | DNA Sequence Determinants Controlling Affinity, Stability and Shape of DNA Complexes Bound by the Nucleoid Protein Fis.
Plos One, 11, 2016
|
|
1ETX
 
 | |
1ETK
 
 | |
1ETY
 
 | |
1ETO
 
 | |
1ETQ
 
 | |
1ETV
 
 | |
1ETW
 
 | |
1F36
 
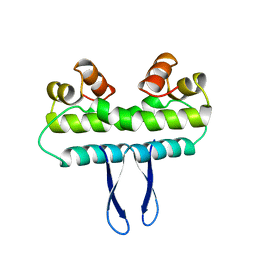 | |
1FIA
 
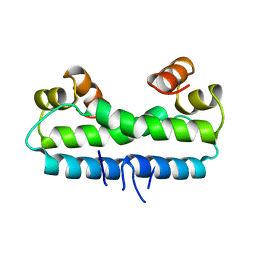 | | CRYSTAL STRUCTURE OF THE FACTOR FOR INVERSION STIMULATION FIS AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Kostrewa, D, Granzin, J, Choe, H.-W, Labahn, J, Saenger, W. | | Deposit date: | 1991-12-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the factor for inversion stimulation FIS at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
1FIP
 
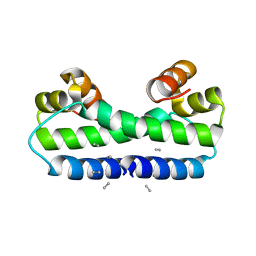 | | THE STRUCTURE OF FIS MUTANT PRO61ALA ILLUSTRATES THAT THE KINK WITHIN THE LONG ALPHA-HELIX IS NOT DUE TO THE PRESENCE OF THE PROLINE RESIDUE | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS), UNKNOWN PEPTIDE, POSSIBLY PART OF THE UNOBSERVED RESIDUES IN ENTITY 1 | | Authors: | Yuan, H.S, Wang, S.S, Yang, W.-Z, Finkel, S.E, Johnson, R.C. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of Fis mutant Pro61Ala illustrates that the kink within the long alpha-helix is not due to the presence of the proline residue.
J.Biol.Chem., 269, 1994
|
|
3IV5
 
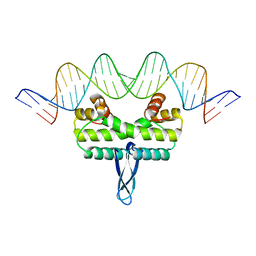 | |
