5AJ9
 
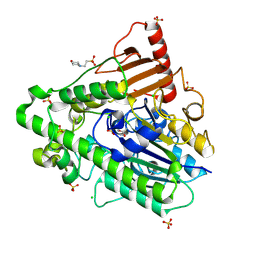 | | G7 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ARYLSULFATASE, CALCIUM ION, ... | | Authors: | Miton, C.M, Fischer, G, Jonas, S, Mohammed, M.F, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8DI0
 
 | |
3LXQ
 
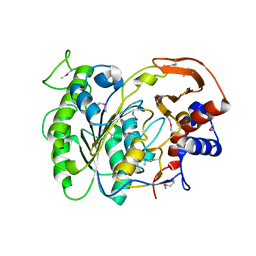 | | The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A | | Descriptor: | CHLORIDE ION, Uncharacterized protein VP1736 | | Authors: | Stein, A.J, Weger, A, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A
To be Published
|
|
3B5Q
 
 | |
3ED4
 
 | | Crystal structure of putative arylsulfatase from escherichia coli | | Descriptor: | ARYLSULFATASE, GLYCEROL, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Gilmore, M, Chang, S, Bain, K, Wasserman, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Arylsulfatase from Escherichia Coli
To be Published
|
|
5WCX
 
 | |
4KAV
 
 | |
5YLF
 
 | | MCR-1 complex with D-glucose | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, beta-D-glucopyranose | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
4KAY
 
 | |
5LRN
 
 | | Structure of mono-zinc MCR-1 in P21 space group | | Descriptor: | GLYCEROL, Phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Hinchliffe, P, Paterson, N.G, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
5LRM
 
 | | Structure of di-zinc MCR-1 in P41212 space group | | Descriptor: | GLYCEROL, ZINC ION, phosphatidylethanolamine transferase Mcr-1 | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
6S20
 
 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33336S-sulf) | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, N-acetylgalactosamine-6-O-sulfatase, ... | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
6S21
 
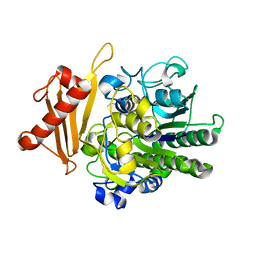 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33494S-sulf) | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Endo-4-O-sulfatase | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
5MX9
 
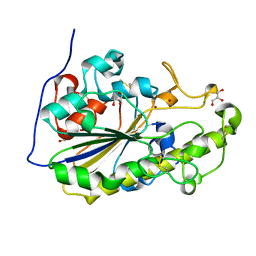 | | High resolution crystal structure of the MCR-2 catalytic domain | | Descriptor: | GLYCEROL, Phosphatidylethanolamine transferase Mcr-2, ZINC ION | | Authors: | Hinchliffe, P, Coates, K, Walsh, T.R, Spencer, J. | | Deposit date: | 2017-01-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 1.12 angstrom resolution crystal structure of the catalytic domain of the plasmid-mediated colistin resistance determinant MCR-2.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6SUT
 
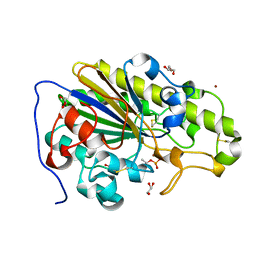 | | Crystal structure of phosphothreonine MCR-2 | | Descriptor: | BROMIDE ION, GLYCEROL, Putative integral membrane protein, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-09-16 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Resistance to the "last resort" antibiotic colistin: a single-zinc mechanism for phosphointermediate formation in MCR enzymes.
Chem.Commun.(Camb.), 56, 2020
|
|
7OQD
 
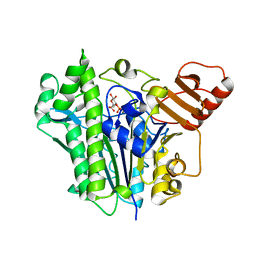 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1636-S1_20) | | Descriptor: | 3-O-sulfo-beta-D-galactopyranose, Arylsulfatase, CALCIUM ION | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2021-06-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
2QZU
 
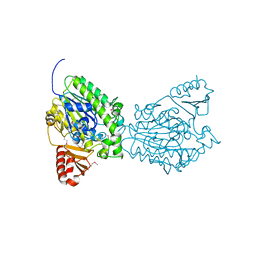 | | Crystal structure of the putative sulfatase yidJ from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR123 | | Descriptor: | Putative sulfatase yidJ | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Wang, D, Cunningham, K, Maglaqui, M, Owens, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-04 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structure of the putative sulfatase yidJ from Bacteroides fragilis.
To be Published
|
|
2VQR
 
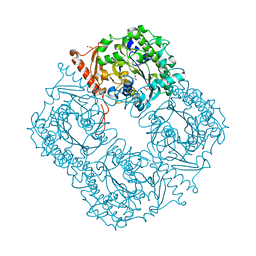 | | Crystal structure of a phosphonate monoester hydrolase from rhizobium leguminosarum: a new member of the alkaline phosphatase superfamily | | Descriptor: | ACETATE ION, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Jonas, S, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2008-03-18 | | Release date: | 2008-09-30 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A New Member of the Alkaline Phosphatase Superfamily with a Formylglycine Nucleophile: Structural and Kinetic Characterisation of a Phosphonate Monoester Hydrolase/Phosphodiesterase from Rhizobium Leguminosarum.
J.Mol.Biol., 384, 2008
|
|
2W5Q
 
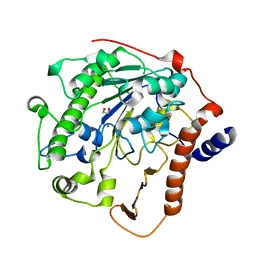 | | Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, PROCESSED GLYCEROL PHOSPHATE LIPOTEICHOIC ACID SYNTHASE | | Authors: | Lu, D, Wormann, M.E, Zhang, X, Scheewind, O, Grundling, A, Freemont, P.S. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Mechanism of Lipoteichoic Acid Synthesis by Staphylococcus Aureus Ltas.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6DGM
 
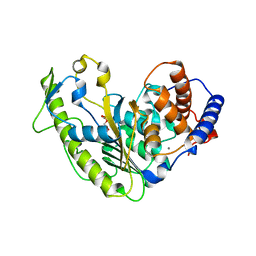 | | Streptococcus pyogenes phosphoglycerol transferase GacH in complex with sn-glycerol-1-phosphate | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Phosphoglycerol transferase GacH, ... | | Authors: | Edgar, R.J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-05-17 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides.
Nat.Chem.Biol., 15, 2019
|
|
7ALL
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT4683-S1_4) | | Descriptor: | Arylsulfatase, CALCIUM ION, IODIDE ION, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Martens, E.C, Basle, A, Cartmell, A. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7ANB
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7AN1
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1636-S1_20) | | Descriptor: | Arylsulfatase, CALCIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-10 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7ANA
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
5K4P
 
 | | Catalytic Domain of MCR-1 phosphoethanolamine transferase | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, sorbitol | | Authors: | Stojanoski, V, Palzkill, T, Prasad, B.V.V, Sankaran, B. | | Deposit date: | 2016-05-21 | | Release date: | 2016-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Structure of the catalytic domain of the colistin resistance enzyme MCR-1.
Bmc Biol., 14, 2016
|
|
