[English] 日本語
 Yorodumi
Yorodumi- EMDB-8301: Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assem... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8301 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Complex Formed by the Iron Donor, the Sulfur Donor, and the Scaffold | |||||||||
 Map data Map data | Human mitochondrial iron-sulfur cluster assembly machinery: grouping of segments corresponding to one half of the full map (EMD-8293) | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of iron ion import across plasma membrane / molybdopterin cofactor metabolic process /  regulation of ferrochelatase activity / Molybdenum cofactor biosynthesis / [4Fe-4S] cluster assembly / regulation of ferrochelatase activity / Molybdenum cofactor biosynthesis / [4Fe-4S] cluster assembly /  proprioception / iron incorporation into metallo-sulfur cluster / positive regulation of lyase activity / positive regulation of succinate dehydrogenase activity / L-cysteine desulfurase complex ...negative regulation of iron ion import across plasma membrane / molybdopterin cofactor metabolic process / proprioception / iron incorporation into metallo-sulfur cluster / positive regulation of lyase activity / positive regulation of succinate dehydrogenase activity / L-cysteine desulfurase complex ...negative regulation of iron ion import across plasma membrane / molybdopterin cofactor metabolic process /  regulation of ferrochelatase activity / Molybdenum cofactor biosynthesis / [4Fe-4S] cluster assembly / regulation of ferrochelatase activity / Molybdenum cofactor biosynthesis / [4Fe-4S] cluster assembly /  proprioception / iron incorporation into metallo-sulfur cluster / positive regulation of lyase activity / positive regulation of succinate dehydrogenase activity / L-cysteine desulfurase complex / Mitochondrial iron-sulfur cluster biogenesis / mitochondrial iron-sulfur cluster assembly complex / positive regulation of aconitate hydratase activity / positive regulation of mitochondrial electron transport, NADH to ubiquinone / iron chaperone activity / proprioception / iron incorporation into metallo-sulfur cluster / positive regulation of lyase activity / positive regulation of succinate dehydrogenase activity / L-cysteine desulfurase complex / Mitochondrial iron-sulfur cluster biogenesis / mitochondrial iron-sulfur cluster assembly complex / positive regulation of aconitate hydratase activity / positive regulation of mitochondrial electron transport, NADH to ubiquinone / iron chaperone activity /  cysteine desulfurase / cysteine desulfurase /  cysteine desulfurase activity / negative regulation of organ growth / iron-sulfur cluster assembly complex / Mo-molybdopterin cofactor biosynthetic process / cysteine desulfurase activity / negative regulation of organ growth / iron-sulfur cluster assembly complex / Mo-molybdopterin cofactor biosynthetic process /  Mitochondrial protein import / [2Fe-2S] cluster assembly / adult walking behavior / Mitochondrial protein import / [2Fe-2S] cluster assembly / adult walking behavior /  oxidative phosphorylation / response to iron ion / embryo development ending in birth or egg hatching / oxidative phosphorylation / response to iron ion / embryo development ending in birth or egg hatching /  iron-sulfur cluster assembly / heme biosynthetic process / muscle cell cellular homeostasis / negative regulation of multicellular organism growth / organ growth / positive regulation of catalytic activity / iron-sulfur cluster assembly / heme biosynthetic process / muscle cell cellular homeostasis / negative regulation of multicellular organism growth / organ growth / positive regulation of catalytic activity /  ferroxidase / ferroxidase /  iron-sulfur cluster binding / negative regulation of release of cytochrome c from mitochondria / iron-sulfur cluster binding / negative regulation of release of cytochrome c from mitochondria /  ferroxidase activity / protein autoprocessing / mitochondrion organization / ferroxidase activity / protein autoprocessing / mitochondrion organization /  ferric iron binding / ferric iron binding /  ferrous iron binding / 2 iron, 2 sulfur cluster binding / cellular response to hydrogen peroxide / ferrous iron binding / 2 iron, 2 sulfur cluster binding / cellular response to hydrogen peroxide /  pyridoxal phosphate binding / Maturation of replicase proteins / iron ion transport / positive regulation of cell growth / intracellular iron ion homeostasis / molecular adaptor activity / pyridoxal phosphate binding / Maturation of replicase proteins / iron ion transport / positive regulation of cell growth / intracellular iron ion homeostasis / molecular adaptor activity /  mitochondrial matrix / iron ion binding / mitochondrial matrix / iron ion binding /  centrosome / positive regulation of cell population proliferation / negative regulation of apoptotic process / protein homodimerization activity / centrosome / positive regulation of cell population proliferation / negative regulation of apoptotic process / protein homodimerization activity /  mitochondrion / zinc ion binding / mitochondrion / zinc ion binding /  nucleoplasm / nucleoplasm /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
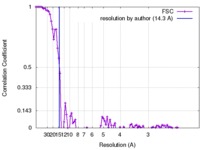
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 14.3 Å negative staining / Resolution: 14.3 Å | |||||||||
 Authors Authors | Gakh O / Ranatunga W / Smith DY / Ahlgren EC / Al-Karadaghi S / Thompson JR / Isaya G | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2016 Journal: J Biol Chem / Year: 2016Title: Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assembly Machinery. Authors: Oleksandr Gakh / Wasantha Ranatunga / Douglas Y Smith / Eva-Christina Ahlgren / Salam Al-Karadaghi / James R Thompson / Grazia Isaya /   Abstract: Fe-S clusters, essential cofactors needed for the activity of many different enzymes, are assembled by conserved protein machineries inside bacteria and mitochondria. As the architecture of the human ...Fe-S clusters, essential cofactors needed for the activity of many different enzymes, are assembled by conserved protein machineries inside bacteria and mitochondria. As the architecture of the human machinery remains undefined, we co-expressed in Escherichia coli the following four proteins involved in the initial step of Fe-S cluster synthesis: FXN (iron donor); [NFS1]·[ISD11] (sulfur donor); and ISCU (scaffold upon which new clusters are assembled). We purified a stable, active complex consisting of all four proteins with 1:1:1:1 stoichiometry. Using negative staining transmission EM and single particle analysis, we obtained a three-dimensional model of the complex with ∼14 Å resolution. Molecular dynamics flexible fitting of protein structures docked into the EM map of the model revealed a [FXN]·[NFS1]·[ISD11]·[ISCU] complex, consistent with the measured 1:1:1:1 stoichiometry of its four components. The complex structure fulfills distance constraints obtained from chemical cross-linking of the complex at multiple recurring interfaces, involving hydrogen bonds, salt bridges, or hydrophobic interactions between conserved residues. The complex consists of a central roughly cubic [FXN]·[ISCU] sub-complex with one symmetric ISCU trimer bound on top of one symmetric FXN trimer at each of its eight vertices. Binding of 12 [NFS1]·[ISD11] sub-complexes to the surface results in a globular macromolecule with a diameter of ∼15 nm and creates 24 Fe-S cluster assembly centers. The organization of each center recapitulates a previously proposed conserved mechanism for sulfur donation from NFS1 to ISCU and reveals, for the first time, a path for iron donation from FXN to ISCU. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8301.map.gz emd_8301.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8301-v30.xml emd-8301-v30.xml emd-8301.xml emd-8301.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8301_fsc.xml emd_8301_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_8301.png emd_8301.png | 85.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8301 http://ftp.pdbj.org/pub/emdb/structures/EMD-8301 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8301 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8301 | HTTPS FTP |
-Related structure data
| Related structure data |  5kz5MC  8293C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8301.map.gz / Format: CCP4 / Size: 11.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8301.map.gz / Format: CCP4 / Size: 11.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human mitochondrial iron-sulfur cluster assembly machinery: grouping of segments corresponding to one half of the full map (EMD-8293) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : NFS1-ISD11-ISCU-FXN
| Entire | Name: NFS1-ISD11-ISCU-FXN |
|---|---|
| Components |
|
-Supramolecule #1: NFS1-ISD11-ISCU-FXN
| Supramolecule | Name: NFS1-ISD11-ISCU-FXN / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant plasmid: pCDF, pET Escherichia coli (E. coli) / Recombinant plasmid: pCDF, pET |
-Macromolecule #1: Cysteine desulfurase, mitochondrial
| Macromolecule | Name: Cysteine desulfurase, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO / EC number:  cysteine desulfurase cysteine desulfurase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.429648 KDa |
| Recombinant expression | Organism:   Escherichia coli #1/H766 (bacteria) Escherichia coli #1/H766 (bacteria) |
| Sequence | String: TPLDPRVLDA MLPYLINYYG NPHSRTHAYG WESEAAMERA RQQVASLIGA DPREIIFTSG ATESNNIAIK GVARFYRSRK KHLITTQTE HKCVLDSCRS LEAEGFQVTY LPVQKSGIID LKELEAAIQP DTSLVSVMTV NNEIGVKQPI AEIGRICSSR K VYFHTDAA ...String: TPLDPRVLDA MLPYLINYYG NPHSRTHAYG WESEAAMERA RQQVASLIGA DPREIIFTSG ATESNNIAIK GVARFYRSRK KHLITTQTE HKCVLDSCRS LEAEGFQVTY LPVQKSGIID LKELEAAIQP DTSLVSVMTV NNEIGVKQPI AEIGRICSSR K VYFHTDAA QAVGKIPLDV NDMKIDLMSI SGHKIYGPKG VGAIYIRRRP RVRVEALQSG GGQERGMRSG TVPTPLVVGL GA ACEVAQQ EMEYDHKRIS KLSERLIQNI MKSLPDVVMN GDPKHHYPGC INLSFAYVEG ESLLMALKDV ALSSGSACTS ASL EPSYVL RAIGTDEDLA HSSIRFGIGR FTTEEEVDYT VEKCIQHVKR LREMSPLWEM VQDGIDLKSI KWTQH |
-Macromolecule #2: Frataxin, mitochondrial
| Macromolecule | Name: Frataxin, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO / EC number:  ferroxidase ferroxidase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.849025 KDa |
| Recombinant expression | Organism:   Escherichia coli #1/H766 (bacteria) Escherichia coli #1/H766 (bacteria) |
| Sequence | String: LRTDIDATCT PRRASSNQRG LNQIWNVKKQ SVYLMNLRKS GTLGHPGSLD ETTYERLAEE TLDSLAEFFE DLADKPYTFE DYDVSFGSG VLTVKLGGDL GTYVINKQTP NKQIWLSSPS SGPKRYDWTG KNWVYSHDGV SLHELLAAEL TKALKTKLDL S SLAYSGKD A |
-Macromolecule #3: Iron-sulfur cluster assembly enzyme ISCU, mitochondrial
| Macromolecule | Name: Iron-sulfur cluster assembly enzyme ISCU, mitochondrial type: protein_or_peptide / ID: 3 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.52558 KDa |
| Recombinant expression | Organism:   Escherichia coli #1/H766 (bacteria) Escherichia coli #1/H766 (bacteria) |
| Sequence | String: GSLDKTSKNV GTGLVGAPAC GDVMKLQIQV DEKGKIVDAR FKTFGCGSAI ASSSLATEWV KGKTVEEALT IKNTDIAKEL CLPPVKLHC SMLAEDAIKA ALADYKLKQE PKKGEAEKK |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: 1% uranyl acetate / Details: 5 and 30 seconds | |||||||||
| Grid | Model: Carbon-coated copper grids, EMS / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Details: DV-502A vacuum evaporator (Denton Vacuum Inc.) | |||||||||
| Details | 50 mM Tris-HCl, pH 8.0, 150 mM NaCl |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.21 µm / Calibrated magnification: 115000 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.21 µm / Nominal magnification: 115000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.21 µm / Nominal magnification: 115000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 466 / Average electron dose: 30.0 e/Å2 |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-5kz5: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)