[English] 日本語
 Yorodumi
Yorodumi- EMDB-8181: Broadly neutralizing antibody PGDM14 in complex with BG505 SOSIP.... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8181 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
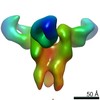
| Title | Broadly neutralizing antibody PGDM14 in complex with BG505 SOSIP.664 Env trimer | |||||||||
 Map data Map data | Broadly neutralizing antibody PGDM14 in complex with BG505 SOSIP.664 Env trimer | |||||||||
 Sample Sample |
| |||||||||
| Biological species |    Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 21.0 Å negative staining / Resolution: 21.0 Å | |||||||||
 Authors Authors | Lee JH / Ward AB | |||||||||
 Citation Citation |  Journal: Immunity / Year: 2016 Journal: Immunity / Year: 2016Title: A Prominent Site of Antibody Vulnerability on HIV Envelope Incorporates a Motif Associated with CCR5 Binding and Its Camouflaging Glycans. Authors: Devin Sok / Matthias Pauthner / Bryan Briney / Jeong Hyun Lee / Karen L Saye-Francisco / Jessica Hsueh / Alejandra Ramos / Khoa M Le / Meaghan Jones / Joseph G Jardine / Raiza Bastidas / ...Authors: Devin Sok / Matthias Pauthner / Bryan Briney / Jeong Hyun Lee / Karen L Saye-Francisco / Jessica Hsueh / Alejandra Ramos / Khoa M Le / Meaghan Jones / Joseph G Jardine / Raiza Bastidas / Anita Sarkar / Chi-Hui Liang / Sachin S Shivatare / Chung-Yi Wu / William R Schief / Chi-Huey Wong / Ian A Wilson / Andrew B Ward / Jiang Zhu / Pascal Poignard / Dennis R Burton /   Abstract: The dense patch of high-mannose-type glycans surrounding the N332 glycan on the HIV envelope glycoprotein (Env) is targeted by multiple broadly neutralizing antibodies (bnAbs). This region is ...The dense patch of high-mannose-type glycans surrounding the N332 glycan on the HIV envelope glycoprotein (Env) is targeted by multiple broadly neutralizing antibodies (bnAbs). This region is relatively conserved, implying functional importance, the origins of which are not well understood. Here we describe the isolation of new bnAbs targeting this region. Examination of these and previously described antibodies to Env revealed that four different bnAb families targeted the (324)GDIR(327) peptide stretch at the base of the gp120 V3 loop and its nearby glycans. We found that this peptide stretch constitutes part of the CCR5 co-receptor binding site, with the high-mannose patch glycans serving to camouflage it from most antibodies. GDIR-glycan bnAbs, in contrast, bound both (324)GDIR(327) peptide residues and high-mannose patch glycans, which enabled broad reactivity against diverse HIV isolates. Thus, as for the CD4 binding site, bnAb effectiveness relies on circumventing the defenses of a critical functional region on Env. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8181.map.gz emd_8181.map.gz | 13.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8181-v30.xml emd-8181-v30.xml emd-8181.xml emd-8181.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8181.png emd_8181.png | 33.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8181 http://ftp.pdbj.org/pub/emdb/structures/EMD-8181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8181 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8181.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8181.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Broadly neutralizing antibody PGDM14 in complex with BG505 SOSIP.664 Env trimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BG505 SOSIP.664 trimer in complex with PGDM14 Fab
| Entire | Name: BG505 SOSIP.664 trimer in complex with PGDM14 Fab |
|---|---|
| Components |
|
-Supramolecule #1: BG505 SOSIP.664 trimer in complex with PGDM14 Fab
| Supramolecule | Name: BG505 SOSIP.664 trimer in complex with PGDM14 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 / Strain: BG505 Human immunodeficiency virus 1 / Strain: BG505 |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK293F / Recombinant plasmid: pPPI4 Homo sapiens (human) / Recombinant cell: HEK293F / Recombinant plasmid: pPPI4 |
-Macromolecule #1: BG505 SOSIP.664 soluble Env trimer
| Macromolecule | Name: BG505 SOSIP.664 soluble Env trimer / type: protein_or_peptide / ID: 1 Details: This soluble Env sequence is derived from HIV-1 isolate BG505. It contains mutations to generate the N332 N-linked glycosylation site (T332N) and has stabilizing mutations A501C, I559P, and ...Details: This soluble Env sequence is derived from HIV-1 isolate BG505. It contains mutations to generate the N332 N-linked glycosylation site (T332N) and has stabilizing mutations A501C, I559P, and T605C. The construct is truncated at residue 664 of GP41. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 / Strain: BG505 Human immunodeficiency virus 1 / Strain: BG505 |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPCV KLTPLCVTLQ CTNVTNNITD DMRGELKNCS FNMTTELRDK KQKVYSLFYR LDVVQINENQ GNRSNNSNKE YRLINCNTSA ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPCV KLTPLCVTLQ CTNVTNNITD DMRGELKNCS FNMTTELRDK KQKVYSLFYR LDVVQINENQ GNRSNNSNKE YRLINCNTSA ITQACPKVSF EPIPIHYCAP AGFAILKCKD KKFNGTGPCP SVSTVQCTHG IKPVVSTQLL LNGSLAEEEV MIRSENITNN AKNILVQFNT PVQINCTRPN NNTRKSIRIG PGQAFYATGD IIGDIRQAHC NVSKATWNET LGKVVKQLRK HFGNNTIIRF ANSSGGDLEV TTHSFNCGGE FFYCNTSGLF NSTWISNTSV QGSNSTGSND SITLPCRIKQ IINMWQRIGQ AMYAPPIQGV IRCVSNITGL ILTRDGGSTN STTETFRPGG GDMRDNWRSE LYKYKVVKIE PLGVAPTRCK RRVVGRRRRR RAVGIGAVFL GFLGAAGSTM GAASMTLTVQ ARNLLSGIVQ QQSNLLRAPE AQQHLLKLTV WGIKQLQARV LAVERYLRDQ QLLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD |
-Macromolecule #2: Heavy chain Fab of broadly neutralizing antibody PGDM14
| Macromolecule | Name: Heavy chain Fab of broadly neutralizing antibody PGDM14 type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVKPGGSLRL SCTASGYYMS WIRQAPGKGL EWIAYISLRN NGYTHYADSV KGRFTMSRDY AKNLMFLQMN SLRVDDTAIY YCARVASGPN EDFYHDTEAA FDIWGQGTPV FVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP VTVSWNSGAL ...String: EVQLVESGGG LVKPGGSLRL SCTASGYYMS WIRQAPGKGL EWIAYISLRN NGYTHYADSV KGRFTMSRDY AKNLMFLQMN SLRVDDTAIY YCARVASGPN EDFYHDTEAA FDIWGQGTPV FVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSCD |
-Macromolecule #3: Light chain of broadly neutralizing antibody PGDM14
| Macromolecule | Name: Light chain of broadly neutralizing antibody PGDM14 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQTTLS SPVTLGQAAS ISCKSSRNLV NSDGNTYLSW LHQR PGQPP SLLIYKISRR FSGVSDRFSG SGAGTDFTLR INRVEAADVG VYYCMQGSHW AWA FGQGTK LHVNRRTVAA PSVFIFPPSD EQLKSGTASV VCLLNNFYPR EAKVQWKVDN ALQSGNSQES ...String: DIVMTQTTLS SPVTLGQAAS ISCKSSRNLV NSDGNTYLSW LHQR PGQPP SLLIYKISRR FSGVSDRFSG SGAGTDFTLR INRVEAADVG VYYCMQGSHW AWA FGQGTK LHVNRRTVAA PSVFIFPPSD EQLKSGTASV VCLLNNFYPR EAKVQWKVDN ALQSGNSQES VTEQDSKDST YSLSSTLTLS KADYEKHKVY ACEVTHQGLS SPVTKSFNRG EC |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: 2% uranyl formate | |||||||||
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 52000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Temperature | Max: 298.0 K |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number grids imaged: 2 / Number real images: 223 / Average electron dose: 25.0 e/Å2 |
- Image processing
Image processing
| Startup model | Type of model: OTHER Details: Common lines model generated from reference-free 2D class averages |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: SPARX / Software - details: sxali3d.py |
| Final reconstruction | Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 21.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPARX / Software - details: sxali3d.py / Number images used: 8493 |
 Movie
Movie Controller
Controller