+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8168 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
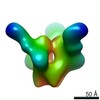
| Title | BF520.1 Fab fragment bound to BG505 T332N SOSIP.664 trimer | |||||||||
 Map data Map data | BF520.1 Fab fragment bound to BG505 T332N SOSIP.664 trimer | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / structural molecule activity / virion attachment to host cell / host cell plasma membrane / virion membrane / identical protein binding / viral envelope / structural molecule activity / virion attachment to host cell / host cell plasma membrane / virion membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /    Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 10.9 Å negative staining / Resolution: 10.9 Å | |||||||||
 Authors Authors | Williams JA / Lee KK | |||||||||
 Citation Citation |  Journal: Cell / Year: 2016 Journal: Cell / Year: 2016Title: HIV-1 Neutralizing Antibodies with Limited Hypermutation from an Infant. Authors: Cassandra A Simonich / Katherine L Williams / Hans P Verkerke / James A Williams / Ruth Nduati / Kelly K Lee / Julie Overbaugh /   Abstract: HIV-1 broadly neutralizing antibodies (bnAbs) develop in a subset of infected adults and exhibit high levels of somatic hypermutation (SHM) due to years of affinity maturation. There is no precedent ...HIV-1 broadly neutralizing antibodies (bnAbs) develop in a subset of infected adults and exhibit high levels of somatic hypermutation (SHM) due to years of affinity maturation. There is no precedent for eliciting highly mutated antibodies by vaccination, nor is it practical to wait years for a desired response. Infants develop broad responses early, which may suggest a more direct path to generating bnAbs. Here, we isolated ten neutralizing antibodies (nAbs) contributing to plasma breadth of an infant at ∼1 year post-infection, including one with cross-clade breadth. The nAbs bind to envelope trimer from the transmitted virus, suggesting that this interaction may have initiated development of the infant nAbs. The infant cross-clade bnAb targets the N332 supersite on envelope but, unlike adult bnAbs targeting this site, lacks indels and has low SHM. The identification of this infant bnAb illustrates that HIV-1-specific neutralization breadth can develop without prolonged affinity maturation and extensive SHM. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8168.map.gz emd_8168.map.gz | 484.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8168-v30.xml emd-8168-v30.xml emd-8168.xml emd-8168.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8168_fsc.xml emd_8168_fsc.xml | 3.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_8168.png emd_8168.png | 60 KB | ||
| Others |  emd_8168_half_map_1.map.gz emd_8168_half_map_1.map.gz emd_8168_half_map_2.map.gz emd_8168_half_map_2.map.gz | 476.9 KB 474.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8168 http://ftp.pdbj.org/pub/emdb/structures/EMD-8168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8168 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8168.map.gz / Format: CCP4 / Size: 2.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8168.map.gz / Format: CCP4 / Size: 2.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BF520.1 Fab fragment bound to BG505 T332N SOSIP.664 trimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.14 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: BF520.1 Fab fragment bound to BG505 T332N SOSIP.664...
| File | emd_8168_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BF520.1 Fab fragment bound to BG505 T332N SOSIP.664 trimer, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: BF520.1 Fab fragment bound to BG505 T332N SOSIP.664...
| File | emd_8168_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BF520.1 Fab fragment bound to BG505 T332N SOSIP.664 trimer, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BF520.1 Fab complexed with BG505.C2 T332N SOSIP.664 trimer
| Entire | Name: BF520.1 Fab complexed with BG505.C2 T332N SOSIP.664 trimer |
|---|---|
| Components |
|
-Supramolecule #1: BF520.1 Fab complexed with BG505.C2 T332N SOSIP.664 trimer
| Supramolecule | Name: BF520.1 Fab complexed with BG505.C2 T332N SOSIP.664 trimer type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: BF520.1 Fab
| Supramolecule | Name: BF520.1 Fab / type: complex / ID: 2 / Parent: 1 Details: Fab domains generated by papain digestion of BF520.1 IgG |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: BG505.C2 T332N SOSIP.664 trimer
| Supramolecule | Name: BG505.C2 T332N SOSIP.664 trimer / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 / Strain: BG505.C2 T332N Human immunodeficiency virus 1 / Strain: BG505.C2 T332N |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK 293F / Recombinant plasmid: pPPI4 Homo sapiens (human) / Recombinant cell: HEK 293F / Recombinant plasmid: pPPI4 |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.020 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Staining | Type: NEGATIVE / Material: methylamine tungstate |
| Grid | Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 52000 Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 24.0 e/Å2 |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z
Z Y
Y X
X



















