[English] 日本語
 Yorodumi
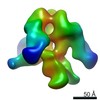
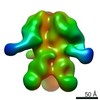
Yorodumi- EMDB-6593: CSF_trimer in a post-fusion or intermediate conformation bound to... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6593 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CSF_trimer in a post-fusion or intermediate conformation bound to the bnAb PGV04 | |||||||||
 Map data Map data | CSF_trimer_post_fusion bound to bnAb PGV04 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  HIV / HIV /  envelope / envelope /  glycoprotein / trimer glycoprotein / trimer | |||||||||
| Biological species |    Human immunodeficiency virus / Human immunodeficiency virus /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 20.0 Å negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Kong L / He L / de Val N / Morris CD / Vora N / Azadnia P / Sok D / Zhou B / Burton DR / Ward AB ...Kong L / He L / de Val N / Morris CD / Vora N / Azadnia P / Sok D / Zhou B / Burton DR / Ward AB / Wilson IA / Zhu J | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2016 Journal: Nat Commun / Year: 2016Title: Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Authors: Leopold Kong / Linling He / Natalia de Val / Nemil Vora / Charles D Morris / Parisa Azadnia / Devin Sok / Bin Zhou / Dennis R Burton / Andrew B Ward / Ian A Wilson / Jiang Zhu /  Abstract: The trimeric HIV-1 envelope glycoprotein (Env) is critical for host immune recognition and neutralization. Despite advances in trimer design, the roots of Env trimer metastability remain elusive. ...The trimeric HIV-1 envelope glycoprotein (Env) is critical for host immune recognition and neutralization. Despite advances in trimer design, the roots of Env trimer metastability remain elusive. Here we investigate the contribution of two Env regions to metastability. First, we computationally redesign a largely disordered bend in heptad region 1 (HR1) of SOSIP trimers that connects the long, central HR1 helix to the fusion peptide, substantially improving the yield of soluble, well-folded trimers. Structural and antigenic analyses of two distinct HR1 redesigns confirm that redesigned Env closely mimics the native, prefusion trimer with a more stable gp41. Next, we replace the cleavage site between gp120 and gp41 with various linkers in the context of an HR1 redesign. Electron microscopy reveals a potential fusion intermediate state for uncleaved trimers containing short but not long linkers. Together, these results outline a general approach for stabilization of Env trimers from diverse HIV-1 strains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6593.map.gz emd_6593.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6593-v30.xml emd-6593-v30.xml emd-6593.xml emd-6593.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6593.gif 400_6593.gif 80_6593.gif 80_6593.gif | 10.9 KB 1.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6593 http://ftp.pdbj.org/pub/emdb/structures/EMD-6593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6593 | HTTPS FTP |
-Related structure data
| Related structure data |  6590C  6591C  6592C  6621C  6622C  6623C  6624C  6625C  6626C  6627C  6628C  5js9C  5jsaC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6593.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6593.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CSF_trimer_post_fusion bound to bnAb PGV04 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CSF trimer post-fusion state or intermediate state bound to PGV04
| Entire | Name: CSF trimer post-fusion state or intermediate state bound to PGV04 |
|---|---|
| Components |
|
-Supramolecule #1000: CSF trimer post-fusion state or intermediate state bound to PGV04
| Supramolecule | Name: CSF trimer post-fusion state or intermediate state bound to PGV04 type: sample / ID: 1000 / Oligomeric state: Trimer bound to three Fabs / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: CSF
| Macromolecule | Name: CSF / type: protein_or_peptide / ID: 1 Details: Heterodimer of gp120 and gp41 assembles into a trimer. Number of copies: 2 / Oligomeric state: trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus Human immunodeficiency virus |
| Molecular weight | Experimental: 570 KDa / Theoretical: 570 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant strain: HEK 293F / Recombinant cell: 293F Homo sapiens (human) / Recombinant strain: HEK 293F / Recombinant cell: 293F |
-Macromolecule #2: PGV04
| Macromolecule | Name: PGV04 / type: protein_or_peptide / ID: 2 / Name.synonym: Fab / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK 293F Homo sapiens (human) / Recombinant cell: HEK 293F |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM Tris-HCl, 150 mM NaCl |
| Staining | Type: NEGATIVE Details: Grids were glow-discharged at 20 mA for 30 seconds and stained with 2% uranyl formate for 40 seconds. |
| Grid | Details: 400 Cu mesh grids |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 46000 Bright-field microscopy / Nominal defocus max: 1.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 46000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 50 |
| Date | Aug 15, 2015 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 119 / Average electron dose: 30.15 e/Å2 |
| Tilt angle min | 0 |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Final two d classification | Number classes: 104 |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: OTHER / Software - Name: EMAN2, sparx / Number images used: 10706 |
 Movie
Movie Controller
Controller