[English] 日本語
 Yorodumi
Yorodumi- EMDB-6373: JRFL Env SOSIP.664 in complex with CD4-binding site hybrid broadl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6373 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | JRFL Env SOSIP.664 in complex with CD4-binding site hybrid broadly neutralizing antibody DRVIA7/VRC01 | |||||||||
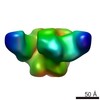
 Map data Map data | Negative-stain reconstruction of DRVIA7/VRC01 hybrid antibody Fab bound to HIV-1 JRFL SOSIP.664 gp140 trimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  HIV-1 / Env / HIV-1 / Env /  broadly-neutralizing antibody / CD4 binding site / SOSIP broadly-neutralizing antibody / CD4 binding site / SOSIP | |||||||||
| Biological species |    Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 22.0 Å negative staining / Resolution: 22.0 Å | |||||||||
 Authors Authors | Ozorowski G / Ward AB | |||||||||
 Citation Citation |  Journal: Immunity / Year: 2016 Journal: Immunity / Year: 2016Title: Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor. Authors: Leopold Kong / Bin Ju / Yajing Chen / Linling He / Li Ren / Jiandong Liu / Kunxue Hong / Bin Su / Zheng Wang / Gabriel Ozorowski / Xiaolin Ji / Yuanzi Hua / Yanli Chen / Marc C Deller / ...Authors: Leopold Kong / Bin Ju / Yajing Chen / Linling He / Li Ren / Jiandong Liu / Kunxue Hong / Bin Su / Zheng Wang / Gabriel Ozorowski / Xiaolin Ji / Yuanzi Hua / Yanli Chen / Marc C Deller / Yanling Hao / Yi Feng / Fernando Garces / Richard Wilson / Kaifan Dai / Sijy O'Dell / Krisha McKee / John R Mascola / Andrew B Ward / Richard T Wyatt / Yuxing Li / Ian A Wilson / Jiang Zhu / Yiming Shao /   Abstract: VRC01-class antibodies neutralize diverse HIV-1 strains by targeting the conserved CD4-binding site. Despite extensive investigations, crucial events in the early stage of VRC01 development remain ...VRC01-class antibodies neutralize diverse HIV-1 strains by targeting the conserved CD4-binding site. Despite extensive investigations, crucial events in the early stage of VRC01 development remain elusive. We demonstrated how VRC01-class antibodies emerged in a Chinese donor by antigen-specific single B cell sorting, structural and functional studies, and longitudinal antibody and virus repertoire analyses. A monoclonal antibody DRVIA7 with modest neutralizing breadth was isolated that displayed a subset of VRC01 signatures. X-ray and EM structures revealed a VRC01-like angle of approach, but less favorable interactions between the DRVIA7 light-chain CDR1 and the N terminus with N276 and V5 glycans of gp120. Although the DRVIA7 lineage was unable to acquire broad neutralization, longitudinal analysis revealed a repertoire-encoded VRC01 light-chain CDR3 signature and VRC01-like neutralizing heavy-chain precursors that rapidly matured within 2 years. Thus, light chain accommodation of the glycan shield should be taken into account in vaccine design targeting this conserved site of vulnerability. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6373.map.gz emd_6373.map.gz | 14 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6373-v30.xml emd-6373-v30.xml emd-6373.xml emd-6373.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6373.png emd_6373.png | 49.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6373 http://ftp.pdbj.org/pub/emdb/structures/EMD-6373 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6373 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6373 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6373.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6373.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative-stain reconstruction of DRVIA7/VRC01 hybrid antibody Fab bound to HIV-1 JRFL SOSIP.664 gp140 trimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Fab of broadly neutralizing antibody hybrid DRVIA7/VRC01 in compl...
| Entire | Name: Fab of broadly neutralizing antibody hybrid DRVIA7/VRC01 in complex with HIV-1 JRFL Env SOSIP.664 trimer |
|---|---|
| Components |
|
-Supramolecule #1000: Fab of broadly neutralizing antibody hybrid DRVIA7/VRC01 in compl...
| Supramolecule | Name: Fab of broadly neutralizing antibody hybrid DRVIA7/VRC01 in complex with HIV-1 JRFL Env SOSIP.664 trimer type: sample / ID: 1000 Details: Incubated Fab with trimer overnight at room temperature prior to placing on grid. Oligomeric state: Trimer of JRFL SOSIP.664 with one Fab bound to each protomer Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: HIV-1 Env SOSIP gp140
| Macromolecule | Name: HIV-1 Env SOSIP gp140 / type: protein_or_peptide / ID: 1 / Name.synonym: SOSIP.664 / Number of copies: 1 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 / Strain: JRFL / synonym: HIV-1 Human immunodeficiency virus 1 / Strain: JRFL / synonym: HIV-1 |
| Molecular weight | Theoretical: 420 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Macromolecule #2: DRVIA7/VRC01 hybrid anti-HIV antibody Fab
| Macromolecule | Name: DRVIA7/VRC01 hybrid anti-HIV antibody Fab / type: protein_or_peptide / ID: 2 / Details: Heavy chain of DRVIA7 and light chain of VRC01 / Number of copies: 3 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 50 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM Tris-HCl, pH 7.4, 150 mM NaCl |
| Staining | Type: NEGATIVE Details: Grids containing adsorbed protein were treated with 3 uL of 2% w/v uranyl formate for 60 seconds. Uranyl formate was then removed with blotting paper. |
| Grid | Details: Glow-discharged CU400 copper grids with carbon support |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Electron beam | Acceleration voltage: 100 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: -1.0 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 52000 Bright-field microscopy / Nominal defocus max: -1.0 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 52000 |
| Specialist optics | Energy filter - Name: FEI |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle min: -50 |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 52,000 magnification. |
| Details | A series of tilts from 0 to 50 degrees in 10 degree increments was collected to increase side views. |
| Date | Aug 22, 2014 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 118 / Average electron dose: 24.44 e/Å2 |
| Tilt angle max | 0 |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: OTHER / Software - Name: EMAN2, Sparx / Number images used: 31789 |
|---|---|
| Details | Particles were selected automatically using DogPicker. Initial 2D class averages were inspected; those displaying complexes of trimer with Fab were subjected another round of classification. Only classes that clearly showed two or three Fabs per trimer were used for reconstruction. |
 Movie
Movie Controller
Controller