[English] 日本語
 Yorodumi
Yorodumi- EMDB-6367: Electron cryo-microscopy 3D reconstruction of an octahedral DNA/A... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6367 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy 3D reconstruction of an octahedral DNA/AuNP hybrid nanoparticle | |||||||||
 Map data Map data | 3D reconstruction of hybrid nanoparticle with an octahedron DNA cage encapsulating a gold nanoparticle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA/AuNP hybrid nanoparticle | |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 24.0 Å cryo EM / Resolution: 24.0 Å | |||||||||
 Authors Authors | Yu G / Yan R / Zhang C / Mao C / Jiang W | |||||||||
 Citation Citation |  Journal: Small / Year: 2015 Journal: Small / Year: 2015Title: Single-Particle Cryo-EM and 3D Reconstruction of Hybrid Nanoparticles with Electron-Dense Components. Authors: Guimei Yu / Rui Yan / Chuan Zhang / Chengde Mao / Wen Jiang /  Abstract: Single-particle cryo-electron microscopy (cryo-EM), accompanied with 3D reconstruction, is a broadly applicable tool for the structural characterization of macromolecules and nanoparticles. Recently, ...Single-particle cryo-electron microscopy (cryo-EM), accompanied with 3D reconstruction, is a broadly applicable tool for the structural characterization of macromolecules and nanoparticles. Recently, the cryo-EM field has pushed the limits of this technique to higher resolutions and samples of smaller molecular mass, however, some samples still present hurdles to this technique. Hybrid particles with electron-dense components, which have been studied using single-particle cryo-EM yet with limited success in 3D reconstruction due to the interference caused by electron-dense elements, constitute one group of such challenging samples. To process such hybrid particles, a masking method is developed in this work to adaptively remove pixels arising from electron-dense portions in individual projection images while maintaining maximal biomass signals for subsequent 2D alignment, 3D reconstruction, and iterative refinements. As demonstrated by the success in 3D reconstruction of an octahedron DNA/gold hybrid particle, which has been previously published without a 3D reconstruction, the devised strategy that combines adaptive masking and standard single-particle 3D reconstruction approach has overcome the hurdle of electron-dense elements interference, and is generally applicable to cryo-EM structural characterization of most, if not all, hybrid nanomaterials with electron-dense components. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6367.map.gz emd_6367.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6367-v30.xml emd-6367-v30.xml emd-6367.xml emd-6367.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
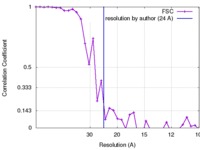
| FSC (resolution estimation) |  emd_6367_fsc.xml emd_6367_fsc.xml | 3.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_6367.png emd_6367.png | 214.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6367 http://ftp.pdbj.org/pub/emdb/structures/EMD-6367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6367 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6367.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6367.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of hybrid nanoparticle with an octahedron DNA cage encapsulating a gold nanoparticle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.98 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Hybrid nanoparticle of octahedral DNA cage encapsulating a gold n...
| Entire | Name: Hybrid nanoparticle of octahedral DNA cage encapsulating a gold nanoparticle |
|---|---|
| Components |
|
-Supramolecule #1000: Hybrid nanoparticle of octahedral DNA cage encapsulating a gold n...
| Supramolecule | Name: Hybrid nanoparticle of octahedral DNA cage encapsulating a gold nanoparticle type: sample / ID: 1000 Oligomeric state: one octahedral DNA cage encapsulating one gold nanoparticle Number unique components: 2 |
|---|
-Macromolecule #1: octahedral DNA nanocage
| Macromolecule | Name: octahedral DNA nanocage / type: dna / ID: 1 / Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #2: gold nanoparticle
| Macromolecule | Name: gold nanoparticle / type: em_label / ID: 2 / Number of copies: 1 |
|---|
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Grid | Details: 400 mesh copper grid with perforated carbon |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal magnification: 66000 Bright-field microscopy / Cs: 2 mm / Nominal magnification: 66000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Date | Jun 5, 2013 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Average electron dose: 20 e/Å2 |
 Movie
Movie Controller
Controller