[English] 日本語
 Yorodumi
Yorodumi- EMDB-6077: Characterization of a broadly neutralizing monoclonal antibody th... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6077 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
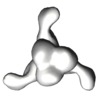
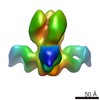
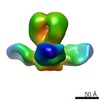
| Title | Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin | |||||||||
 Map data Map data | Negative stain reconstruction of Fab 9H10 bound to hemagglutinin (H3) from Influenza A/Hong Kong/1/1968 (H3N2), initiated with common lines, refined with projection matching against single particles, under C3 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | group 2 influenza A virus /  broadly neutralizing antibody broadly neutralizing antibody | |||||||||
| Biological species |    Influenza A virus / Influenza A virus /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 24.6 Å negative staining / Resolution: 24.6 Å | |||||||||
 Authors Authors | Tan GS / Lee PS / Hoffman RMB / Mazel-Sanchez B / Krammer F / Leon PE / Ward AB / Wilson IA / Palese P | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2014 Journal: J Virol / Year: 2014Title: Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. Authors: Gene S Tan / Peter S Lee / Ryan M B Hoffman / Beryl Mazel-Sanchez / Florian Krammer / Paul E Leon / Andrew B Ward / Ian A Wilson / Peter Palese /  Abstract: Due to continuous changes to its antigenic regions, influenza viruses can evade immune detection and cause a significant amount of morbidity and mortality around the world. Influenza vaccinations can ...Due to continuous changes to its antigenic regions, influenza viruses can evade immune detection and cause a significant amount of morbidity and mortality around the world. Influenza vaccinations can protect against disease but must be annually reformulated to match the current circulating strains. In the development of a broad-spectrum influenza vaccine, the elucidation of conserved epitopes is paramount. To this end, we designed an immunization strategy in mice to boost the humoral response against conserved regions of the hemagglutinin (HA) glycoprotein. Of note, generation and identification of broadly neutralizing antibodies that target group 2 HAs are rare and thus far have yielded only a few monoclonal antibodies (MAbs). Here, we demonstrate that mouse MAb 9H10 has broad and potent in vitro neutralizing activity against H3 and H10 group 2 influenza A subtypes. In the mouse model, MAb 9H10 protects mice against two divergent mouse-adapted H3N2 strains, in both pre- and postexposure administration regimens. In vitro and cell-free assays suggest that MAb 9H10 inhibits viral replication by blocking HA-dependent fusion of the viral and endosomal membranes early in the replication cycle and by disrupting viral particle egress in the late stage of infection. Interestingly, electron microscopy reconstructions of MAb 9H10 bound to the HA reveal that it binds a similar binding footprint to MAbs CR8020 and CR8043. IMPORTANCE: The influenza hemagglutinin is the major antigenic target of the humoral immune response. However, due to continuous antigenic changes that occur on the surface of this glycoprotein, ...IMPORTANCE: The influenza hemagglutinin is the major antigenic target of the humoral immune response. However, due to continuous antigenic changes that occur on the surface of this glycoprotein, influenza viruses can escape the immune system and cause significant disease to the host. Toward the development of broad-spectrum therapeutics and vaccines against influenza virus, elucidation of conserved regions of influenza viruses is crucial. Thus, defining these types of epitopes through the generation and characterization of broadly neutralizing monoclonal antibodies (MAbs) can greatly assist others in highlighting conserved regions of hemagglutinin. Here, we demonstrate that MAb 9H10 that targets the hemagglutinin stalk has broadly neutralizing activity against group 2 influenza A viruses in vitro and in vivo. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6077.map.gz emd_6077.map.gz | 959.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6077-v30.xml emd-6077-v30.xml emd-6077.xml emd-6077.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6077.png emd_6077.png | 79.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6077 http://ftp.pdbj.org/pub/emdb/structures/EMD-6077 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6077 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6077 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6077.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6077.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain reconstruction of Fab 9H10 bound to hemagglutinin (H3) from Influenza A/Hong Kong/1/1968 (H3N2), initiated with common lines, refined with projection matching against single particles, under C3 symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Fab 9H10 bound to hemagglutinin (H3) from Influenza A/Hong Kong/1...
| Entire | Name: Fab 9H10 bound to hemagglutinin (H3) from Influenza A/Hong Kong/1/1968 (H3N2) |
|---|---|
| Components |
|
-Supramolecule #1000: Fab 9H10 bound to hemagglutinin (H3) from Influenza A/Hong Kong/1...
| Supramolecule | Name: Fab 9H10 bound to hemagglutinin (H3) from Influenza A/Hong Kong/1/1968 (H3N2) type: sample / ID: 1000 Oligomeric state: one hemagglutinin homotrimer binds to three 9H10 Fabs Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: hemagglutinin
| Macromolecule | Name: hemagglutinin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Oligomeric state: trimer / Recombinant expression: Yes / Database: NCBI |
|---|---|
| Source (natural) | Organism:    Influenza A virus / Strain: A/Hong Kong/1/1968 (H3N2) / synonym: flu virus Influenza A virus / Strain: A/Hong Kong/1/1968 (H3N2) / synonym: flu virus |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #2: Fab of monoclonal antibody 9H10
| Macromolecule | Name: Fab of monoclonal antibody 9H10 / type: protein_or_peptide / ID: 2 / Name.synonym: antigen-binding fragment / Number of copies: 3 / Oligomeric state: monomer / Recombinant expression: Yes / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) / Strain: BALB/c / synonym: house mouse Mus musculus (house mouse) / Strain: BALB/c / synonym: house mouse |
| Molecular weight | Theoretical: 50 KDa |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 150 mM NaCl, 20 mM Tris |
| Staining | Type: NEGATIVE Details: Protein adsorbed for 20 seconds, blotted, stained with 2% uranyl formate for 20 seconds, and blotted again. |
| Grid | Details: 400 mesh copper grid coated with nitrocellulose and a thin layer of carbon |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 Bright-field microscopy / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle min: -55 |
| Temperature | Average: 298 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism monitored/corrected using continuously acquired FFTs at 52000X. |
| Date | Feb 1, 2014 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 283 |
| Tilt angle max | 0 |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 24.6 Å / Resolution method: OTHER / Software - Name: EMAN2, Sparx / Number images used: 4791 |
|---|---|
| Details | Initial model determined by common lines from reference-free class averages. Final model refined against single particles using projection matching. |
 Movie
Movie Controller
Controller