[English] 日本語
 Yorodumi
Yorodumi- EMDB-6003: Cryo-electron tomography of full-length glycoprotein from Ebola v... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6003 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
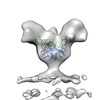
| Title | Cryo-electron tomography of full-length glycoprotein from Ebola virus-like particles | |||||||||
 Map data Map data | Molecular structure of Ebola VLP full-length glycoprotein trimer, including the mucin-like domain | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Ebola / Ebola /  glycoprotein / mucin-like domain glycoprotein / mucin-like domain | |||||||||
| Biological species |   Zaire ebolavirus Zaire ebolavirus | |||||||||
| Method | subtomogram averaging /  cryo EM cryo EM | |||||||||
 Authors Authors | Tran EEH / Simmons JA / Bartesaghi A / Shoemaker CJ / Nelson E / White JM / Subramaniam S | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2014 Journal: J Virol / Year: 2014Title: Spatial localization of the Ebola virus glycoprotein mucin-like domain determined by cryo-electron tomography. Authors: Erin E H Tran / James A Simmons / Alberto Bartesaghi / Charles J Shoemaker / Elizabeth Nelson / Judith M White / Sriram Subramaniam /  Abstract: The Ebola virus glycoprotein mucin-like domain (MLD) is implicated in Ebola virus cell entry and immune evasion. Using cryo-electron tomography of Ebola virus-like particles, we determined a three- ...The Ebola virus glycoprotein mucin-like domain (MLD) is implicated in Ebola virus cell entry and immune evasion. Using cryo-electron tomography of Ebola virus-like particles, we determined a three-dimensional structure for the full-length glycoprotein in a near-native state and compared it to that of a glycoprotein lacking the MLD. Our results, which show that the MLD is located at the apex and the sides of each glycoprotein monomer, provide a structural template for analysis of MLD function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6003.map.gz emd_6003.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6003-v30.xml emd-6003-v30.xml emd-6003.xml emd-6003.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6003.png emd_6003.png | 259 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6003 http://ftp.pdbj.org/pub/emdb/structures/EMD-6003 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6003 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6003 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6003.map.gz / Format: CCP4 / Size: 1.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6003.map.gz / Format: CCP4 / Size: 1.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Molecular structure of Ebola VLP full-length glycoprotein trimer, including the mucin-like domain | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Molecular structure of Ebola VLP full-length glycoprotein trimer
| Entire | Name: Molecular structure of Ebola VLP full-length glycoprotein trimer |
|---|---|
| Components |
|
-Supramolecule #1000: Molecular structure of Ebola VLP full-length glycoprotein trimer
| Supramolecule | Name: Molecular structure of Ebola VLP full-length glycoprotein trimer type: sample / ID: 1000 / Oligomeric state: trimer / Number unique components: 1 |
|---|
-Macromolecule #1: Envelope glycoprotein
| Macromolecule | Name: Envelope glycoprotein / type: protein_or_peptide / ID: 1 Details: Envelope glycoproteins present on the surface of intact virus-like particles Number of copies: 3 / Oligomeric state: trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Zaire ebolavirus / synonym: Ebola Zaire ebolavirus / synonym: Ebola |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK 293T Homo sapiens (human) / Recombinant cell: HEK 293TRecombinant plasmid: pVP40, pBeta-Lactamase-VP40, pmCherry-VP40, pEbola Zaire GP |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 130 mM NaCl, 20 mM HEPES, 10% sucrose |
|---|---|
| Grid | Details: 200 mesh Quantifoil Multi-A |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 77 K / Instrument: FEI VITROBOT MARK III Method: Blot for 6 seconds at 22 degrees C, 100% humidity, blot offset -2, plunge into an ethane slurry cooled by liquid nitrogen. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 34000 Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 34000 |
| Specialist optics | Energy filter - Name: GATAN GIF / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Temperature | Average: 81 K |
| Date | Mar 5, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 37 / Average electron dose: 150 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Software - Name:  IMOD / Number subtomograms used: 5298 IMOD / Number subtomograms used: 5298 |
|---|---|
| Details | Subtomogram density was selected using an automatic selection program. |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: I / Chain - #1 - Chain ID: K / Chain - #2 - Chain ID: M / Chain - #3 - Chain ID: O |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: rigid body, automated fitting procedures |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller