[English] 日本語
 Yorodumi
Yorodumi- EMDB-5330: Cryo-electron tomography reveals novel interactions and doublet-s... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5330 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
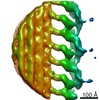
| Title | Cryo-electron tomography reveals novel interactions and doublet-specific structures in the I1 dynein | |||||||||
 Map data Map data | This is a subtomogram average of the I1 inner dynein complex in wild type Chlamydomonas flagella | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  axoneme / axoneme /  molecular motor / motility regulation / molecular motor / motility regulation /  flagella flagella | |||||||||
| Biological species |   Chlamydomonas reinhardtii (plant) Chlamydomonas reinhardtii (plant) | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 39.0 Å cryo EM / Resolution: 39.0 Å | |||||||||
 Authors Authors | Heuser T / Barber CF / Lin J / Krell J / Rebesco M / Porter ME / Nicastro D | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2012 Journal: Proc Natl Acad Sci U S A / Year: 2012Title: Cryoelectron tomography reveals doublet-specific structures and unique interactions in the I1 dynein. Authors: Thomas Heuser / Cynthia F Barber / Jianfeng Lin / Jeremy Krell / Matthew Rebesco / Mary E Porter / Daniela Nicastro /  Abstract: Cilia and flagella are highly conserved motile and sensory organelles in eukaryotes, and defects in ciliary assembly and motility cause many ciliopathies. The two-headed I1 inner arm dynein is a ...Cilia and flagella are highly conserved motile and sensory organelles in eukaryotes, and defects in ciliary assembly and motility cause many ciliopathies. The two-headed I1 inner arm dynein is a critical regulator of ciliary and flagellar beating. To understand I1 architecture and function better, we analyzed the 3D structure and composition of the I1 dynein in Chlamydomonas axonemes by cryoelectron tomography and subtomogram averaging. Our data revealed several connections from the I1 dynein to neighboring structures that are likely to be important for assembly and/or regulation, including a tether linking one I1 motor domain to the doublet microtubule and doublet-specific differences potentially contributing to the asymmetrical distribution of dynein activity required for ciliary beating. We also imaged three I1 mutants and analyzed their polypeptide composition using 2D gel-based proteomics. Structural and biochemical comparisons revealed the likely location of the regulatory IC138 phosphoprotein and its associated subcomplex. Overall, our studies demonstrate that I1 dynein is connected to multiple structures within the axoneme, and therefore ideally positioned to integrate signals that regulate ciliary motility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5330.map.gz emd_5330.map.gz | 284.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5330-v30.xml emd-5330-v30.xml emd-5330.xml emd-5330.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5330_1.tif emd_5330_1.tif | 184.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5330 http://ftp.pdbj.org/pub/emdb/structures/EMD-5330 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5330 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5330 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5330.map.gz / Format: CCP4 / Size: 336.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5330.map.gz / Format: CCP4 / Size: 336.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a subtomogram average of the I1 inner dynein complex in wild type Chlamydomonas flagella | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 9.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-electron tomography and subtomographic average (750 axonemal...
| Entire | Name: Cryo-electron tomography and subtomographic average (750 axonemal repeats) of isolated axonemes of wild type Chlamydomonas (CC 125, 137c), I1 dynein complex (dynein f) is bound to the doublet ...Name: Cryo-electron tomography and subtomographic average (750 axonemal repeats) of isolated axonemes of wild type Chlamydomonas (CC 125, 137c), I1 dynein complex (dynein f) is bound to the doublet microtubule and is connected to neighboring structures. |
|---|---|
| Components |
|
-Supramolecule #1000: Cryo-electron tomography and subtomographic average (750 axonemal...
| Supramolecule | Name: Cryo-electron tomography and subtomographic average (750 axonemal repeats) of isolated axonemes of wild type Chlamydomonas (CC 125, 137c), I1 dynein complex (dynein f) is bound to the doublet ...Name: Cryo-electron tomography and subtomographic average (750 axonemal repeats) of isolated axonemes of wild type Chlamydomonas (CC 125, 137c), I1 dynein complex (dynein f) is bound to the doublet microtubule and is connected to neighboring structures. type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: I1 dynein complex
| Supramolecule | Name: I1 dynein complex / type: organelle_or_cellular_component / ID: 1 / Name.synonym: dynein f / Number of copies: 2 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Chlamydomonas reinhardtii (plant) / Strain: CC-125, 137c / synonym: unicellular green algae / Cell: Chlamydomonas reinhardtii / Organelle: eukaryotic flagella Chlamydomonas reinhardtii (plant) / Strain: CC-125, 137c / synonym: unicellular green algae / Cell: Chlamydomonas reinhardtii / Organelle: eukaryotic flagella |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 10 mM HEPES, pH 7.4, 25 mM NaCl, 4 mM MgSO4, 1 mM EGTA, 0.1 mM EDTA |
| Grid | Details: Quantifoil holey carbon grids Cu 200 mesh R2/2 |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 100 K / Instrument: HOMEMADE PLUNGER / Method: front-side blotting for 2-3 seconds |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 8.0 µm / Nominal defocus min: 6.0 µm / Nominal magnification: 13500 Bright-field microscopy / Nominal defocus max: 8.0 µm / Nominal defocus min: 6.0 µm / Nominal magnification: 13500 |
| Specialist optics | Energy filter - Name: GATAN postcolumn filter GIF / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Temperature | Average: 80 K |
| Date | Feb 20, 2004 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (2k x 2k) / Average electron dose: 100 e/Å2 |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 39.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name:  IMOD IMODDetails: Final maps were calculated by averaging 750 particles from 5 tomograms |
|---|---|
| Details | 750 axonemal repeats (96 nm long) from 5 tomograms (reconstructed using fiducial alignment and weighted backprojection, IMOD software, Kremer et al. 1996) were aligned and averaged using the PEET software (bio3d.colorado.edu, Nicastro et al. 2006). Average number of tilts used in the 3D reconstructions: 80. Average tomographic tilt angle increment: 1.5. |
 Movie
Movie Controller
Controller