+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3410 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM of nanoscale DNA assemblies | |||||||||
 Map data Map data | Reconstruction of a DNA octahedron object | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Scaffolded DNA origami | |||||||||
| Biological species |   Enterobacteria phage M13 (virus) / synthetic construct (others) Enterobacteria phage M13 (virus) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 25.0 Å cryo EM / Resolution: 25.0 Å | |||||||||
 Authors Authors | Zhang K / Chiu W / Veneziano R / Ratanalert S / Zhang F / Yan H / Bathe M | |||||||||
 Citation Citation |  Journal: Science / Year: 2016 Journal: Science / Year: 2016Title: Designer nanoscale DNA assemblies programmed from the top down. Authors: Rémi Veneziano / Sakul Ratanalert / Kaiming Zhang / Fei Zhang / Hao Yan / Wah Chiu / Mark Bathe /  Abstract: Scaffolded DNA origami is a versatile means of synthesizing complex molecular architectures. However, the approach is limited by the need to forward-design specific Watson-Crick base pairing manually ...Scaffolded DNA origami is a versatile means of synthesizing complex molecular architectures. However, the approach is limited by the need to forward-design specific Watson-Crick base pairing manually for any given target structure. Here, we report a general, top-down strategy to design nearly arbitrary DNA architectures autonomously based only on target shape. Objects are represented as closed surfaces rendered as polyhedral networks of parallel DNA duplexes, which enables complete DNA scaffold routing with a spanning tree algorithm. The asymmetric polymerase chain reaction is applied to produce stable, monodisperse assemblies with custom scaffold length and sequence that are verified structurally in three dimensions to be high fidelity by single-particle cryo-electron microscopy. Their long-term stability in serum and low-salt buffer confirms their utility for biological as well as nonbiological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3410.map.gz emd_3410.map.gz | 9.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3410-v30.xml emd-3410-v30.xml emd-3410.xml emd-3410.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-3410.png EMD-3410.png | 171.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3410 http://ftp.pdbj.org/pub/emdb/structures/EMD-3410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3410 | HTTPS FTP |
-Related structure data
| Related structure data | 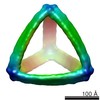 3408C  3409C  3411C  3412C  3413C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10067 (Title: Designer nanoscale DNA assemblies programmed from the top down EMPIAR-10067 (Title: Designer nanoscale DNA assemblies programmed from the top downData size: 7.3 / Data #1: DNA octahedron [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3410.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3410.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of a DNA octahedron object | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.51 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DNA octahedron, 52-bp edge length
| Entire | Name: DNA octahedron, 52-bp edge length |
|---|---|
| Components |
|
-Supramolecule #1000: DNA octahedron, 52-bp edge length
| Supramolecule | Name: DNA octahedron, 52-bp edge length / type: sample / ID: 1000 Details: Monodisperse and pure sample were prepared by overnight folding reaction and purified using centrifugal filter (MWCO 100KDa) Oligomeric state: monomer / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 940 KDa / Theoretical: 940 KDa / Method: Calculation |
-Macromolecule #1: M13mp18 phage genome segment
| Macromolecule | Name: M13mp18 phage genome segment / type: dna / ID: 1 / Name.synonym: Scaffold strand / Details: amplified using assymetric PCR / Classification: DNA / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:   Enterobacteria phage M13 (virus) / Strain: M13mp18 / synonym: M13 phage Enterobacteria phage M13 (virus) / Strain: M13mp18 / synonym: M13 phage |
| Molecular weight | Experimental: 520 KDa / Theoretical: 520 KDa |
| Sequence | String: GCGACGATTT ACAGAAGCAA GGTTATTCAC TCACATATAT TGATTTATGT ACTGTTTCCA TTAAAAAAGG TAATTCAAAT GAAATTGTTA AATGTAATTA ATTTTGTTTT CTTGATGTTT GTTTCATCAT CTTCTTTTGC TCAGGTAATT GAAATGAATA ATTCGCCTCT ...String: GCGACGATTT ACAGAAGCAA GGTTATTCAC TCACATATAT TGATTTATGT ACTGTTTCCA TTAAAAAAGG TAATTCAAAT GAAATTGTTA AATGTAATTA ATTTTGTTTT CTTGATGTTT GTTTCATCAT CTTCTTTTGC TCAGGTAATT GAAATGAATA ATTCGCCTCT GCGCGATTTT GTAACTTGGT ATTCAAAGCA ATCAGGCGAA TCCGTTATTG TTTCTCCCGA TGTAAAAGGT ACTGTTACTG TATATTCATC TGACGTTAAA CCTGAAAATC TACGCAATTT CTTTATTTCT GTTTTACGTG CTAATAATTT TGATATGGTT GGTTCAATTC CTTCCATAAT TCAGAAGTAT AATCCAAACA ATCAGGATTA TATTGATGAA TTGCCATCAT CTGATAATCA GGAATATGAT GATAATTCCG CTCCTTCTGG TGGTTTCTTT GTTCCGCAAA ATGATAATGT TACTCAAACT TTTAAAATTA ATAACGTTCG GGCAAAGGAT TTAATACGAG TTGTCGAATT GTTTGTAAAG TCTAATACTT CTAAATCCTC AAATGTATTA TCTATTGACG GCTCTAATCT ATTAGTTGTT AGTGCACCTA AAGATATTTT AGATAACCTT CCTCAATTCC TTTCTACTGT TGATTTGCCA ACTGACCAGA TATTGATTGA GGGTTTGATA TTTGAGGTTC AGCAAGGTGA TGCTTTAGAT TTTTCATTTG CTGCTGGCTC TCAGCGTGGC ACTGTTGCAG GCGGTGTTAA TACTGACCGC CTCACCTCTG TTTTATCTTC TGCTGGTGGT TCGTTCGGTA TTTTTAATGG CGATGTTTTA GGGCTATCAG TTCGCGCATT AAAGACTAAT AGCCATTCAA AAATATTGTC TGTGCCACGT ATTCTTACGC TTTCAGGTCA GAAGGGTTCT ATCTCTGTTG GCCAGAATGT CCCTTTTATT ACTGGTCGTG TGACTGGTGA ATCTGCCAAT GTAAATAATC CATTTCAGAC GATTGAGCGT CAAAATGTAG GTATTTCCAT GAGCGTTTTT CCTGTTGCAA TGGCTGGCGG TAATATTGTT CTGGATATTA CCAGCAAGGC CGATAGTTTG AGTTCTTCTA CTCAGGCAAG TGATGTTATT ACTAATCAAA GAAGTATTGC TACAACGGTT AATTTGCGTG ATGGACAGAC TCTTTTACTC GGTGGCCTCA CTGATTATAA AAACACTTCT CAAGATTCTG GCGTACCGTT CCTGTCTAAA ATCCCTTTAA TCGGCCTCCT GTTTAGCTCC CGCTCTGATT CCAACGAGGA AAGCACGTTA TACGTGCTCG TCAAAGCAAC CATAGTACGC GCCCTGTAGC GGCGCATTAA GCGCGGCGGG TGTGGTGGTT ACGCGCAGCG TGACCGCTAC ACTTGCCAGC GCCCTAGCGC CCGCTCCTTT CGCTTTCTTC CCTTCCTTTC TCGCCACGTT CGCCGGCTTT CCCCGTCAAG CTCTAAATCG GGGGCTCCCT TTAGGGTTCC GATTTAGTGC TTTACGGCAC CTCGACCCCA AAAAACTTGA TTTGGGTGAT GGTTCACGTA GTGGGCCATC GCCCTGATAG ACGGTTTTTC GCCCTT |
-Macromolecule #2: Synthetic DNA oligonucleotides
| Macromolecule | Name: Synthetic DNA oligonucleotides / type: dna / ID: 2 / Name.synonym: Staple strand Details: TAGAGCCGTCATTTTTATAGATAATACTGGCCAACAGTTTTTAGATAGAACC CCAGTAATAATTTAGAAGTATTAGACTTTACCAGTCACACGA AAGGGACATTCATTTGAGGA TTAATGGAAACTTTTTAGTACATAAAAATAACGGATTTTTTTCGCCTGATTG ...Details: TAGAGCCGTCATTTTTATAGATAATACTGGCCAACAGTTTTTAGATAGAACC CCAGTAATAATTTAGAAGTATTAGACTTTACCAGTCACACGA AAGGGACATTCATTTGAGGA TTAATGGAAACTTTTTAGTACATAAAAATAACGGATTTTTTTCGCCTGATTG CCTTTTACATTGAGTGAATAACCAAAGGGATCAGTAACAGTA CGGGAGAAACTCAATATATG TTTAGACAGGATTTTTACGGTACGCCTATTTACATTGTTTTTGCAGATTCAC AAGTGTTTTTGCTCAATCGTCTGAAATGGATAGAATCTTGAG ATAATCAGTGACATTTTGAC AAACAATTCGATTTTTCAACTCGTATGTTTAACGTCATTTTTGATGAATATA CCCGAACGTTAGAAATTGCGTAGATTTTCAGTAAATCCTTTG ATTAATTTTAACAGAAATAA CACAGACAATATTTTTTTTTTGAATGATTGAGGAAGGTTTTTTTATCTAAAA CAAATCAACATTTAATGCGCGAACTGATAGCTGGTCAGTTGG GTAGAAAGGAGCTATTAGTC AGGCGAATTATTTTTTTCATTTCAATTCAAGAAAACATTTTTAAATTAATTA AACAAACATACCTGAGCAAAAGAAACAGTGC CACGCTGATTAACACCGCCTGCAGATGATGA CCTAAAACATCTTTTTGCCATTAAAAACAGAGGTGAGTTTTTGCGGTCAGTA CAAACTATATCACTTGCCTGAGTACCAGCAG AAGATAAAATACCGAACGAACCAGAAGAACT GAGCCAGCAGCTTTTTAAATGAAAAAATCAAACCCTCTTTTTAATCAATATC AATATAATATTATCAGATGATGGCTGCTGAA CCTCAAATTCTAAAGCATCACCTAATTCATC CGGCCTTGCTGTTTTTGTAATATCCAAAAACGCTCATTTTTTGGAAATACCT CAACAGGAGAACAATATTACCGCCCGTAAGA ATACGTGGCTTCTGACCTGAAAGAGCCATTG AAAGTTTGAGTTTTTTAACATTATCAGGAATTATCATTTTTTCATATTCCTG GAAGGAGCTTTTGCGGAACAAAGATAACAAC TAATAGATTATCTTTAGGTGCACAACCACCA AGGCCACCGAGTTTTTTAAAAGAGTCACTTCTTTGATTTTTTTAGTAATAAC GTAGCAATTGTCCATCACGCAAATTTTGAAT TACCTTTTCATTTAACAATTTCATAACCGTT CCTGATTGTTTTTTTTGGATTATACTTCAAAATTATTTTTTTAGCACGTAAA CAACCATATCTGAATTATGGAAGGTACAAAA TCGCGCAGCTTTGAATACCAAGTAATTGAAC Classification: DNA / Structure: SINGLE STRANDED / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Experimental: 420 KDa / Theoretical: 420 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 40 mM Tris-HCl, 20 mM acetic acid, 2 mM EDTA |
| Grid | Details: glow-discharged 200-mesh R1.2/1.3 Quantifoil grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Method: Blot for 1.5 second before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 25000 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 25000 |
| Specialist optics | Energy filter - Name: JEOL |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 80,000 times magnification |
| Details | 5k * 4k |
| Date | Sep 19, 2015 |
| Image recording | Category: CCD / Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Number real images: 100 / Average electron dose: 63 e/Å2 Details: Every image is the average of 60 frames recorded by the direct electron detecto |
- Image processing
Image processing
| CTF correction | Details: each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: O (octahedral ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER / Software - Name: EMAN2 / Number images used: 2678 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER / Software - Name: EMAN2 / Number images used: 2678 |
| Details | The particles were selected manually using EMAN2 |
 Movie
Movie Controller
Controller







