[English] 日本語
 Yorodumi
Yorodumi- EMDB-2625: Negative stain reconstruction of 8ANC195 Fab in complex with BG50... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2625 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
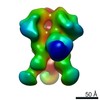
| Title | Negative stain reconstruction of 8ANC195 Fab in complex with BG505 SOSIP.664 trimer | |||||||||
 Map data Map data | Reconstruction of 8ANC195 Fab bound to soluble Env trimer BG505.SOSIP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  HIV-1 / broadly neutralizing antibodies / bnAb / 8ANC195 / SOSIP HIV-1 / broadly neutralizing antibodies / bnAb / 8ANC195 / SOSIP | |||||||||
| Biological species |    Human Immunodeficiency Virus-1 / Human Immunodeficiency Virus-1 /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 18.7 Å negative staining / Resolution: 18.7 Å | |||||||||
 Authors Authors | Lee JH / Scharf L / Bjorkman PJ / Ward AB | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2014 Journal: Cell Rep / Year: 2014Title: Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Authors: Louise Scharf / Johannes F Scheid / Jeong Hyun Lee / Anthony P West / Courtney Chen / Han Gao / Priyanthi N P Gnanapragasam / René Mares / Michael S Seaman / Andrew B Ward / Michel C ...Authors: Louise Scharf / Johannes F Scheid / Jeong Hyun Lee / Anthony P West / Courtney Chen / Han Gao / Priyanthi N P Gnanapragasam / René Mares / Michael S Seaman / Andrew B Ward / Michel C Nussenzweig / Pamela J Bjorkman /  Abstract: Broadly neutralizing antibodies (bNAbs) to HIV-1 envelope glycoprotein (Env) can prevent infection in animal models. Characterized bNAb targets, although key to vaccine and therapeutic strategies, ...Broadly neutralizing antibodies (bNAbs) to HIV-1 envelope glycoprotein (Env) can prevent infection in animal models. Characterized bNAb targets, although key to vaccine and therapeutic strategies, are currently limited. We defined a new site of vulnerability by solving structures of bNAb 8ANC195 complexed with monomeric gp120 by X-ray crystallography and trimeric Env by electron microscopy. The site includes portions of gp41 and N-linked glycans adjacent to the CD4-binding site on gp120, making 8ANC195 the first donor-derived anti-HIV-1 bNAb with an epitope spanning both Env subunits. Rather than penetrating the glycan shield by using a single variable-region CDR loop, 8ANC195 inserted its entire heavy-chain variable domain into a gap to form a large interface with gp120 glycans and regions of the gp120 inner domain not contacted by other bNAbs. By isolating additional 8ANC195 clonal variants, we identified a more potent variant, which may be valuable for therapeutic approaches using bNAb combinations with nonoverlapping epitopes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2625.map.gz emd_2625.map.gz | 13.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2625-v30.xml emd-2625-v30.xml emd-2625.xml emd-2625.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2625.png emd_2625.png | 133 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2625 http://ftp.pdbj.org/pub/emdb/structures/EMD-2625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2625 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2625.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2625.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of 8ANC195 Fab bound to soluble Env trimer BG505.SOSIP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 8ANC195 Fab in complex with BG505 SOSIP.664 gp140 trimer
| Entire | Name: 8ANC195 Fab in complex with BG505 SOSIP.664 gp140 trimer |
|---|---|
| Components |
|
-Supramolecule #1000: 8ANC195 Fab in complex with BG505 SOSIP.664 gp140 trimer
| Supramolecule | Name: 8ANC195 Fab in complex with BG505 SOSIP.664 gp140 trimer type: sample / ID: 1000 / Oligomeric state: One Fab binds per gp140 monomer / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: Soluble HIV-1 Envelope glycoprotein
| Macromolecule | Name: Soluble HIV-1 Envelope glycoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: BG505 SOSIP.664 / Number of copies: 1 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Human Immunodeficiency Virus-1 / Strain: BG505 / synonym: HIV-1 Human Immunodeficiency Virus-1 / Strain: BG505 / synonym: HIV-1 |
| Molecular weight | Theoretical: 350 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK293T Homo sapiens (human) / Recombinant cell: HEK293T |
-Macromolecule #2: Fab 8ANC195
| Macromolecule | Name: Fab 8ANC195 / type: protein_or_peptide / ID: 2 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK293T Homo sapiens (human) / Recombinant cell: HEK293T |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 150 mM NaCl, 50 mM Tris |
| Staining | Type: NEGATIVE Details: Grids with adsorbed sample stained with 2% UF for 30 seconds |
| Grid | Details: 400 mesh Cu with thin carbon support, glow discharged |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 52000 Bright-field microscopy / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Temperature | Max: 293 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism corrected at 52,000x mag |
| Details | Data collected from 0 to -40 degrees in 10 degree increments |
| Date | May 13, 2013 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 195 / Average electron dose: 25 e/Å2 / Details: Data collected on CCD |
| Tilt angle min | 0 |
- Image processing
Image processing
| CTF correction | Details: not corrected |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.7 Å / Resolution method: OTHER / Software - Name: EMAN2, Sparx / Number images used: 11637 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.7 Å / Resolution method: OTHER / Software - Name: EMAN2, Sparx / Number images used: 11637 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: E / Chain - #3 - Chain ID: F / Chain - #4 - Chain ID: I / Chain - #5 - Chain ID: J |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The models were fit using the Fit function in Chimera. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - #0 - Chain ID: G / Chain - #1 - Chain ID: H / Chain - #2 - Chain ID: L |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | This structure was fit by either superimposing the gp120 onto gp120 of the SOSIP trimer, or by using the Fit function in Chimera. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller