[English] 日本語
 Yorodumi
Yorodumi- EMDB-2197: Characterization of the insertase for beta-barrel proteins of the... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2197 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Characterization of the insertase for beta-barrel proteins of the outer mitochondrial membrane. 3-D reconstruction of the TOB complex | |||||||||
 Map data Map data | 3D reconstruction of TOB complexes isolated using the 9xHis tag on the Tob38 subunit. The TOB complex is a hetero trimer containing one copy each of Tob37, Tob38 and Tob37. Total mol. Wgt. Is 140kDa. This sample contains a mixture of three isoforms of Tob55 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TOB/SAM complex /  beta-barrel proteins / Tob55/Sam50 / mitochondria outer membrane beta-barrel proteins / Tob55/Sam50 / mitochondria outer membrane | |||||||||
| Biological species |   Neurospora crassa (fungus) Neurospora crassa (fungus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 15.0 Å cryo EM / Resolution: 15.0 Å | |||||||||
 Authors Authors | Klein A / Israel L / Lackey SWK / Nargang FE / Imhof A / Baumeister W / Neupert W / Thomas DR | |||||||||
 Citation Citation |  Journal: J Cell Biol / Year: 2012 Journal: J Cell Biol / Year: 2012Title: Characterization of the insertase for β-barrel proteins of the outer mitochondrial membrane. Authors: Astrid Klein / Lars Israel / Sebastian W K Lackey / Frank E Nargang / Axel Imhof / Wolfgang Baumeister / Walter Neupert / Dennis R Thomas /  Abstract: The TOB-SAM complex is an essential component of the mitochondrial outer membrane that mediates the insertion of β-barrel precursor proteins into the membrane. We report here its isolation and ...The TOB-SAM complex is an essential component of the mitochondrial outer membrane that mediates the insertion of β-barrel precursor proteins into the membrane. We report here its isolation and determine its size, composition, and structural organization. The complex from Neurospora crassa was composed of Tob55-Sam50, Tob38-Sam35, and Tob37-Sam37 in a stoichiometry of 1:1:1 and had a molecular mass of 140 kD. A very minor fraction of the purified complex was associated with one Mdm10 protein. Using molecular homology modeling for Tob55 and cryoelectron microscopy reconstructions of the TOB complex, we present a model of the TOB-SAM complex that integrates biochemical and structural data. We discuss our results and the structural model in the context of a possible mechanism of the TOB insertase. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2197.map.gz emd_2197.map.gz | 9.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2197-v30.xml emd-2197-v30.xml emd-2197.xml emd-2197.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2197.jpg EMD-2197.jpg | 22.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2197 http://ftp.pdbj.org/pub/emdb/structures/EMD-2197 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2197 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2197 | HTTPS FTP |
-Related structure data
| Related structure data |  2195C 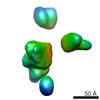 2196C  2200C  2201C  2202C  2203C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2197.map.gz / Format: CCP4 / Size: 10.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2197.map.gz / Format: CCP4 / Size: 10.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of TOB complexes isolated using the 9xHis tag on the Tob38 subunit. The TOB complex is a hetero trimer containing one copy each of Tob37, Tob38 and Tob37. Total mol. Wgt. Is 140kDa. This sample contains a mixture of three isoforms of Tob55 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.78 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TOB complex mitochondrial outer membrane complex required for ins...
| Entire | Name: TOB complex mitochondrial outer membrane complex required for inserting beta-barrel proteins into the outer membrane. |
|---|---|
| Components |
|
-Supramolecule #1000: TOB complex mitochondrial outer membrane complex required for ins...
| Supramolecule | Name: TOB complex mitochondrial outer membrane complex required for inserting beta-barrel proteins into the outer membrane. type: sample / ID: 1000 / Details: The complexes were monodisperse. Oligomeric state: heterotrimer with one subunit each Tob37,Tob38 and Tob55 Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 140 KDa / Theoretical: 140 KDa Method: Blue native gel electrophoresis and Isotope dilution mass spectroscopy analysis of bands isolated from BNGE gels. |
-Macromolecule #1: Tob37
| Macromolecule | Name: Tob37 / type: protein_or_peptide / ID: 1 Details: The TOB complex contains one copy each of T0b37, Tob38 and Tob55 Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Neurospora crassa (fungus) / Strain: Tob38HT / Organelle: mitochondria / Location in cell: outer membrane Neurospora crassa (fungus) / Strain: Tob38HT / Organelle: mitochondria / Location in cell: outer membrane |
| Molecular weight | Experimental: 48.6 KDa / Theoretical: 48.6 KDa |
-Macromolecule #2: Tob38
| Macromolecule | Name: Tob38 / type: protein_or_peptide / ID: 2 Details: The TOB complex contains one copy each of T0b37, Tob38 and Tob55 Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Neurospora crassa (fungus) / Strain: Tob38HT / Organelle: mitochondria / Location in cell: outer membrane Neurospora crassa (fungus) / Strain: Tob38HT / Organelle: mitochondria / Location in cell: outer membrane |
| Molecular weight | Experimental: 37.3 KDa / Theoretical: 37.3 KDa |
-Macromolecule #3: Tob55
| Macromolecule | Name: Tob55 / type: protein_or_peptide / ID: 3 Details: The TOB complex contains one copy each of T0b37, Tob38 and Tob55 Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Neurospora crassa (fungus) / Strain: Tob38HT / Organelle: mitochondria / Location in cell: outer membrane Neurospora crassa (fungus) / Strain: Tob38HT / Organelle: mitochondria / Location in cell: outer membrane |
| Molecular weight | Experimental: 54.7 KDa / Theoretical: 54.7 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8.5 Details: 1mM PMSF, 0.08%(v/v) Triton X-100, 50 mM HEPES pH 8.5 |
| Grid | Details: lacey carbon films on 200 mesh Molybdenum grids |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 120 K / Instrument: HOMEMADE PLUNGER Method: blot for 4-5 seconds before plunging with whatman filter paper #1 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 84270 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: -3.7 µm / Nominal defocus min: -0.5 µm / Nominal magnification: 62000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: -3.7 µm / Nominal defocus min: -0.5 µm / Nominal magnification: 62000 |
| Sample stage | Specimen holder: Gatan 656 side entry holder / Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Max: 95 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected using the live FFT at imaging magnification. |
| Date | May 13, 2010 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (4k x 4k) / Number real images: 318 / Average electron dose: 20 e/Å2 Details: Images collected using TOM_acquisition software. 318 good micrographs were CTF corrected for phase. 104,000 particles were automatically selected. In the end 43500 were included in the final reconstruction. |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Phase and astigmatism correction applied to each micrograph |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 15.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER, TOM_toolbox Details: Final maps were reconstructed from images that had stable alignment parameters over the last 4 rounds of refinement. Stable was defined by absolute accumulated changes in theta and psi of ...Details: Final maps were reconstructed from images that had stable alignment parameters over the last 4 rounds of refinement. Stable was defined by absolute accumulated changes in theta and psi of the projection matched of less than 10 degrees. Number images used: 1 |
 Movie
Movie Controller
Controller




