+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2176 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
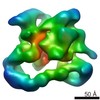
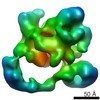
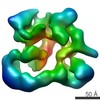
| Title | Negative stain electron microscopy of a CSN complex | |||||||||
 Map data Map data | CSN | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Cop9 Signalosome Cop9 Signalosome | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 25.0 Å negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Enchev RI / Scott DC / da Fonseca P / Schreiber A / Monda JK / Schulman BA / Peter M / Morris EP | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2012 Journal: Cell Rep / Year: 2012Title: Structural basis for a reciprocal regulation between SCF and CSN. Authors: Radoslav I Enchev / Daniel C Scott / Paula C A da Fonseca / Anne Schreiber / Julie K Monda / Brenda A Schulman / Matthias Peter / Edward P Morris /  Abstract: Skp1-Cul1-Fbox (SCF) E3 ligases are activated by ligation to the ubiquitin-like protein Nedd8, which is reversed by the deneddylating Cop9 signalosome (CSN). However, CSN also promotes SCF substrate ...Skp1-Cul1-Fbox (SCF) E3 ligases are activated by ligation to the ubiquitin-like protein Nedd8, which is reversed by the deneddylating Cop9 signalosome (CSN). However, CSN also promotes SCF substrate turnover through unknown mechanisms. Through biochemical and electron microscopy analyses, we determined molecular models of CSN complexes with SCF(Skp2/Cks1) and SCF(Fbw7) and found that CSN occludes both SCF functional sites-the catalytic Rbx1-Cul1 C-terminal domain and the substrate receptor. Indeed, CSN binding prevents SCF interactions with E2 enzymes and a ubiquitination substrate, and it inhibits SCF-catalyzed ubiquitin chain formation independent of deneddylation. Importantly, CSN prevents neddylation of the bound cullin, unless binding of a ubiquitination substrate triggers SCF dissociation and neddylation. Taken together, the results provide a model for how reciprocal regulation sensitizes CSN to the SCF assembly state and inhibits a catalytically competent SCF until a ubiquitination substrate drives its own degradation by displacing CSN, thereby promoting cullin neddylation and substrate ubiquitination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2176.map.gz emd_2176.map.gz | 3.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2176-v30.xml emd-2176-v30.xml emd-2176.xml emd-2176.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2176.jpg EMD-2176.jpg | 19.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2176 http://ftp.pdbj.org/pub/emdb/structures/EMD-2176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2176 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2176.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2176.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CSN | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.47 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CSN
| Entire | Name: CSN |
|---|---|
| Components |
|
-Supramolecule #1000: CSN
| Supramolecule | Name: CSN / type: sample / ID: 1000 Details: The sample was purified over affinity pull-down and Superose6 size exclusion chromatography Oligomeric state: one CSN monomer / Number unique components: 8 |
|---|---|
| Molecular weight | Theoretical: 320 KDa |
-Macromolecule #1: Csn1
| Macromolecule | Name: Csn1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 53 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac |
-Macromolecule #2: Csn2
| Macromolecule | Name: Csn2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 52 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac |
-Macromolecule #3: Csn3
| Macromolecule | Name: Csn3 / type: protein_or_peptide / ID: 3 / Details: N-terminal StrepII2x tag / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 48 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac |
-Macromolecule #4: Csn4
| Macromolecule | Name: Csn4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 46 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac |
-Macromolecule #5: Csn5
| Macromolecule | Name: Csn5 / type: protein_or_peptide / ID: 5 / Details: N-terminal His6 tag / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 40 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac |
-Macromolecule #6: Csn6
| Macromolecule | Name: Csn6 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 36 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac |
-Macromolecule #7: Csn7b
| Macromolecule | Name: Csn7b / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 30 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac |
-Macromolecule #8: Csn8
| Macromolecule | Name: Csn8 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 23 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac Trichoplusia ni (cabbage looper) / Recombinant plasmid: recombinant baculovirus/ MultiBac |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.8 Details: 15 mM HEPES pH 7.8, 150 mM NaCl, 1% glycerol and 1 mM DTT |
| Staining | Type: NEGATIVE Details: Quantifoil grids (R2/2 Cu 400 mesh) coated with thin carbon floated from mica were glow-discharged for 30 seconds at 50 mA and 0.2 mbar vacuum. 3 ul purified samples were applied for 1 min ...Details: Quantifoil grids (R2/2 Cu 400 mesh) coated with thin carbon floated from mica were glow-discharged for 30 seconds at 50 mA and 0.2 mbar vacuum. 3 ul purified samples were applied for 1 min to the grids. Following two brief buffer washes, the grids were stained with 2% uranyl acetate, gently blotted using filter paper and air-dried. |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 50000 Bright-field microscopy / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Date | Jan 10, 2011 |
| Image recording | Category: CCD / Film or detector model: GENERIC TVIPS (4k x 4k) / Average electron dose: 100 e/Å2 / Details: data was collected on a CCD camera |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMAGIC, Spider, EMAN, in-house / Number images used: 5165 |
|---|---|
| Details | see publication |
 Movie
Movie Controller
Controller