[English] 日本語
 Yorodumi
Yorodumi- PDB-3qem: Crystal structure of amino terminal domains of the NMDA receptor ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3qem | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of amino terminal domains of the NMDA receptor subunit GluN1 and GluN2B in complex with Ro 25-6981 | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  TRANSPORT PROTEIN / TRANSPORT PROTEIN /  ion channel / ion channel /  NMDA receptor / NMDA receptor /  allosteric modulation / allosteric modulation /  phenylethanolamine / phenylethanolamine /  N-Glycosylation / N-Glycosylation /  extracellular / extracellular /  transmembrane transmembrane | |||||||||
| Function / homology |  Function and homology information Function and homology informationneurotransmitter receptor activity involved in regulation of postsynaptic membrane potential / cellular response to curcumin / cellular response to corticosterone stimulus / cellular response to magnesium starvation / NMDA selective glutamate receptor signaling pathway / regulation of postsynaptic cytosolic calcium ion concentration / sensory organ development /  sensitization / EPHB-mediated forward signaling / neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration ...neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential / cellular response to curcumin / cellular response to corticosterone stimulus / cellular response to magnesium starvation / NMDA selective glutamate receptor signaling pathway / regulation of postsynaptic cytosolic calcium ion concentration / sensory organ development / sensitization / EPHB-mediated forward signaling / neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration ...neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential / cellular response to curcumin / cellular response to corticosterone stimulus / cellular response to magnesium starvation / NMDA selective glutamate receptor signaling pathway / regulation of postsynaptic cytosolic calcium ion concentration / sensory organ development /  sensitization / EPHB-mediated forward signaling / neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration / Assembly and cell surface presentation of NMDA receptors / regulation of protein kinase A signaling / sensitization / EPHB-mediated forward signaling / neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration / Assembly and cell surface presentation of NMDA receptors / regulation of protein kinase A signaling /  dendritic branch / dendritic branch /  apical dendrite / response to other organism / fear response / response to methylmercury / positive regulation of cysteine-type endopeptidase activity / glutamate-gated calcium ion channel activity / cellular response to dsRNA / response to carbohydrate / regulation of monoatomic cation transmembrane transport / negative regulation of dendritic spine maintenance / apical dendrite / response to other organism / fear response / response to methylmercury / positive regulation of cysteine-type endopeptidase activity / glutamate-gated calcium ion channel activity / cellular response to dsRNA / response to carbohydrate / regulation of monoatomic cation transmembrane transport / negative regulation of dendritic spine maintenance /  interleukin-1 receptor binding / cellular response to lipid / positive regulation of glutamate secretion / interleukin-1 receptor binding / cellular response to lipid / positive regulation of glutamate secretion /  NMDA glutamate receptor activity / response to growth hormone / NMDA selective glutamate receptor complex / RAF/MAP kinase cascade / Synaptic adhesion-like molecules / parallel fiber to Purkinje cell synapse / calcium ion transmembrane import into cytosol / response to manganese ion / protein heterotetramerization / NMDA glutamate receptor activity / response to growth hormone / NMDA selective glutamate receptor complex / RAF/MAP kinase cascade / Synaptic adhesion-like molecules / parallel fiber to Purkinje cell synapse / calcium ion transmembrane import into cytosol / response to manganese ion / protein heterotetramerization /  glutamate binding / glutamate binding /  action potential / action potential /  glycine binding / response to zinc ion / glycine binding / response to zinc ion /  heterocyclic compound binding / receptor clustering / suckling behavior / heterocyclic compound binding / receptor clustering / suckling behavior /  startle response / behavioral response to pain / regulation of neuronal synaptic plasticity / response to amine / monoatomic cation transmembrane transport / startle response / behavioral response to pain / regulation of neuronal synaptic plasticity / response to amine / monoatomic cation transmembrane transport /  regulation of MAPK cascade / regulation of MAPK cascade /  small molecule binding / small molecule binding /  associative learning / response to magnesium ion / positive regulation of excitatory postsynaptic potential / monoatomic cation transport / extracellularly glutamate-gated ion channel activity / cellular response to organic cyclic compound / Unblocking of NMDA receptors, glutamate binding and activation / neuron development / regulation of postsynaptic membrane potential / behavioral fear response / multicellular organismal response to stress / D2 dopamine receptor binding / cellular response to manganese ion / associative learning / response to magnesium ion / positive regulation of excitatory postsynaptic potential / monoatomic cation transport / extracellularly glutamate-gated ion channel activity / cellular response to organic cyclic compound / Unblocking of NMDA receptors, glutamate binding and activation / neuron development / regulation of postsynaptic membrane potential / behavioral fear response / multicellular organismal response to stress / D2 dopamine receptor binding / cellular response to manganese ion /  long-term memory / detection of mechanical stimulus involved in sensory perception of pain / positive regulation of synaptic transmission / response to electrical stimulus / response to mechanical stimulus / glutamate-gated receptor activity / long-term memory / detection of mechanical stimulus involved in sensory perception of pain / positive regulation of synaptic transmission / response to electrical stimulus / response to mechanical stimulus / glutamate-gated receptor activity /  synaptic cleft / presynaptic active zone membrane / response to fungicide / cellular response to forskolin / monoatomic cation channel activity / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / response to amphetamine / response to organonitrogen compound / synaptic cleft / presynaptic active zone membrane / response to fungicide / cellular response to forskolin / monoatomic cation channel activity / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / response to amphetamine / response to organonitrogen compound /  ionotropic glutamate receptor binding / ionotropic glutamate receptor binding /  excitatory postsynaptic potential / hippocampal mossy fiber to CA3 synapse / ionotropic glutamate receptor signaling pathway / excitatory postsynaptic potential / hippocampal mossy fiber to CA3 synapse / ionotropic glutamate receptor signaling pathway /  cell adhesion molecule binding / positive regulation of synaptic transmission, glutamatergic / response to cocaine / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / cell adhesion molecule binding / positive regulation of synaptic transmission, glutamatergic / response to cocaine / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential /  synaptic membrane / synaptic membrane /  synaptic transmission, glutamatergic / synaptic transmission, glutamatergic /  learning / long-term synaptic potentiation / response to cytokine / hippocampus development / cellular response to amino acid stimulus / postsynaptic density membrane / regulation of long-term neuronal synaptic plasticity / learning / long-term synaptic potentiation / response to cytokine / hippocampus development / cellular response to amino acid stimulus / postsynaptic density membrane / regulation of long-term neuronal synaptic plasticity /  regulation of synaptic plasticity / regulation of synaptic plasticity /  calcium channel activity / calcium channel activity /  terminal bouton / response to organic cyclic compound / terminal bouton / response to organic cyclic compound /  memory / response to toxic substance / cerebral cortex development memory / response to toxic substance / cerebral cortex developmentSimilarity search - Function | |||||||||
| Biological species |  Xenopus laevis (African clawed frog) Xenopus laevis (African clawed frog)  Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.003 Å MOLECULAR REPLACEMENT / Resolution: 3.003 Å | |||||||||
 Authors Authors | Karakas, E. / Simorowski, N. / Furukawa, H. | |||||||||
 Citation Citation |  Journal: Nature / Year: 2011 Journal: Nature / Year: 2011Title: Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Authors: Karakas, E. / Simorowski, N. / Furukawa, H. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3qem.cif.gz 3qem.cif.gz | 528.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3qem.ent.gz pdb3qem.ent.gz | 428.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3qem.json.gz 3qem.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qe/3qem https://data.pdbj.org/pub/pdb/validation_reports/qe/3qem ftp://data.pdbj.org/pub/pdb/validation_reports/qe/3qem ftp://data.pdbj.org/pub/pdb/validation_reports/qe/3qem | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3qekSC  3qelC  3jpwS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 4 molecules ACBD
| #1: Protein |  NMDA receptor NMDA receptorMass: 42932.055 Da / Num. of mol.: 2 / Fragment: Amino Terminal Domain, residues 23-405 / Mutation: N61Q, N371Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Xenopus laevis (African clawed frog) / Gene: grin1, NR1 / Production host: Xenopus laevis (African clawed frog) / Gene: grin1, NR1 / Production host:   Trichoplusia ni (cabbage looper) / References: UniProt: Q91977, UniProt: A0A1L8F5J9*PLUS Trichoplusia ni (cabbage looper) / References: UniProt: Q91977, UniProt: A0A1L8F5J9*PLUS#2: Protein | Mass: 41367.902 Da / Num. of mol.: 2 / Fragment: Amino Terminal Domain, residues 31-394 / Mutation: N348D Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rattus norvegicus (Norway rat) / Gene: Grin2b / Production host: Rattus norvegicus (Norway rat) / Gene: Grin2b / Production host:   Trichoplusia ni (cabbage looper) / References: UniProt: Q00960 Trichoplusia ni (cabbage looper) / References: UniProt: Q00960 |
|---|
-Sugars , 2 types, 6 molecules 
| #3: Polysaccharide | alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2- ...alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose / Mass: 910.823 Da / Num. of mol.: 1 / Mass: 910.823 Da / Num. of mol.: 1Source method: isolated from a genetically manipulated source |
|---|---|
| #4: Sugar | ChemComp-NAG /  N-Acetylglucosamine N-Acetylglucosamine |
-Non-polymers , 2 types, 4 molecules 
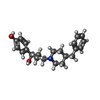

| #5: Chemical | | #6: Chemical | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.15 Å3/Da / Density % sol: 60.97 % |
|---|---|
Crystal grow | Temperature: 290 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: 3.0-3.5M NaFormate, 0.1 M HEPES pH 7.5 , VAPOR DIFFUSION, HANGING DROP, temperature 290K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X29A / Wavelength: 1.075 Å / Beamline: X29A / Wavelength: 1.075 Å | ||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Sep 18, 2010 | ||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Si 111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 1.075 Å / Relative weight: 1 : 1.075 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 3→50 Å / Num. obs: 42325 / % possible obs: 99.9 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -3 / Rmerge(I) obs: 0.09 / Rsym value: 0.09 / Net I/σ(I): 13.8 | ||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3JPW, 3QEK Resolution: 3.003→29.941 Å / SU ML: 0.36 / σ(F): 0 / Phase error: 26.58 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 1.06 Å / VDW probe radii: 1.3 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 47.96 Å2 / ksol: 0.296 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.003→29.941 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj






