[English] 日本語
 Yorodumi
Yorodumi- PDB-8wpz: Cryo-ET structure of RuBisCO at 3.9 angstroms from Synechococcus ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8wpz | ||||||
|---|---|---|---|---|---|---|---|
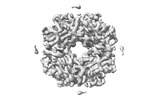
| Title | Cryo-ET structure of RuBisCO at 3.9 angstroms from Synechococcus elongatus PCC 7942 | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  PHOTOSYNTHESIS / PHOTOSYNTHESIS /  carboxysome / carboxysome /  RuBisCO / cryo-et RuBisCO / cryo-et | ||||||
| Function / homology |  Function and homology information Function and homology information carboxysome / carboxysome /  photorespiration / photorespiration /  ribulose-bisphosphate carboxylase / ribulose-bisphosphate carboxylase /  ribulose-bisphosphate carboxylase activity / reductive pentose-phosphate cycle / ribulose-bisphosphate carboxylase activity / reductive pentose-phosphate cycle /  monooxygenase activity / magnesium ion binding monooxygenase activity / magnesium ion bindingSimilarity search - Function | ||||||
| Biological species |   Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) | ||||||
| Method |  ELECTRON MICROSCOPY / subtomogram averaging / ELECTRON MICROSCOPY / subtomogram averaging /  cryo EM / Resolution: 3.9 Å cryo EM / Resolution: 3.9 Å | ||||||
 Authors Authors | Kong, W.W. / Jiang, Y.L. / Zhou, C.Z. | ||||||
| Funding support |  China, 1items China, 1items
| ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-electron tomography reveals the packaging pattern of RuBisCOs in Synechococcus beta-carboxysome Authors: Kong, W.W. / Zhu, Y. / Zhao, H.R. / Du, K. / Zhou, R.Q. / Li, B. / Yang, F. / Hou, P. / Chen, Y. / Sun, F. / Jiang, Y.L. / Zhou, C.Z. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8wpz.cif.gz 8wpz.cif.gz | 756.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8wpz.ent.gz pdb8wpz.ent.gz | 635.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8wpz.json.gz 8wpz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wp/8wpz https://data.pdbj.org/pub/pdb/validation_reports/wp/8wpz ftp://data.pdbj.org/pub/pdb/validation_reports/wp/8wpz ftp://data.pdbj.org/pub/pdb/validation_reports/wp/8wpz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  37727MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein |  RuBisCO / RuBisCO small subunit RuBisCO / RuBisCO small subunitMass: 13349.196 Da / Num. of mol.: 8 / Source method: isolated from a natural source Source: (natural)   Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria)References: UniProt: Q31NB2 #2: Protein | Mass: 52516.605 Da / Num. of mol.: 8 / Source method: isolated from a natural source Source: (natural)   Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria)References: UniProt: Q31NB3,  ribulose-bisphosphate carboxylase ribulose-bisphosphate carboxylase |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: subtomogram averaging |
- Sample preparation
Sample preparation
| Component | Name: RuBisCO / Type: COMPLEX / Entity ID: all / Source: NATURAL / Type: COMPLEX / Entity ID: all / Source: NATURAL |
|---|---|
| Source (natural) | Organism:   Synechococcus elongatus (strain ATCC 33912 / PCC 7942 / FACHB-805) (bacteria) Synechococcus elongatus (strain ATCC 33912 / PCC 7942 / FACHB-805) (bacteria) |
| Details of virus | Empty: NO / Enveloped: NO / Isolate: OTHER / Type: VIRION |
| Natural host | Organism: Nostoc sp. PCC 7120 = FACHB-418 |
| Buffer solution | pH: 8 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 300 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3000 nm / Nominal defocus min: 1000 nm Bright-field microscopy / Nominal defocus max: 3000 nm / Nominal defocus min: 1000 nm |
| Image recording | Electron dose: 3.5 e/Å2 / Avg electron dose per subtomogram: 145 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| EM software | Name: RELION / Version: 3.1 / Category: 3D reconstruction | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING ONLY | ||||||||||||||||||||||||
3D reconstruction | Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 2700 / Symmetry type: POINT | ||||||||||||||||||||||||
| EM volume selection | Num. of tomograms: 88 / Num. of volumes extracted: 29000 | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj
