[English] 日本語
 Yorodumi
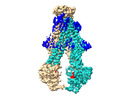
Yorodumi- PDB-8t3k: Heterodimeric ABC transporter BmrCD in the inward-facing conforma... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8t3k | ||||||
|---|---|---|---|---|---|---|---|
| Title | Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to ATP: BmrCD_IF-ATP2 | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  TRANSLOCASE / TRANSLOCASE /  transporter / transporter /  complex / complex /  lipids / lipids /  MEMBRANE PROTEIN MEMBRANE PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationATPase-coupled lipid transmembrane transporter activity / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / transmembrane transport / response to antibiotic /  ATP hydrolysis activity / ATP hydrolysis activity /  ATP binding / ATP binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Bacillus subtilis subsp. subtilis str. 168 (bacteria) Bacillus subtilis subsp. subtilis str. 168 (bacteria) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.33 Å cryo EM / Resolution: 3.33 Å | ||||||
 Authors Authors | Tang, Q. / Mchaourab, H. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Asymmetric conformations and lipid interactions shape the ATP-coupled cycle of a heterodimeric ABC transporter. Authors: Qingyu Tang / Matt Sinclair / Hale S Hasdemir / Richard A Stein / Erkan Karakas / Emad Tajkhorshid / Hassane S Mchaourab /  Abstract: Here we used cryo-electron microscopy (cryo-EM), double electron-electron resonance spectroscopy (DEER), and molecular dynamics (MD) simulations, to capture and characterize ATP- and substrate-bound ...Here we used cryo-electron microscopy (cryo-EM), double electron-electron resonance spectroscopy (DEER), and molecular dynamics (MD) simulations, to capture and characterize ATP- and substrate-bound inward-facing (IF) and occluded (OC) conformational states of the heterodimeric ATP binding cassette (ABC) multidrug exporter BmrCD in lipid nanodiscs. Supported by DEER analysis, the structures reveal that ATP-powered isomerization entails changes in the relative symmetry of the BmrC and BmrD subunits that propagates from the transmembrane domain to the nucleotide binding domain. The structures uncover asymmetric substrate and Mg binding which we hypothesize are required for triggering ATP hydrolysis preferentially in one of the nucleotide-binding sites. MD simulations demonstrate that multiple lipid molecules differentially bind the IF versus the OC conformation thus establishing that lipid interactions modulate BmrCD energy landscape. Our findings are framed in a model that highlights the role of asymmetric conformations in the ATP-coupled transport with general implications to the mechanism of ABC transporters. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8t3k.cif.gz 8t3k.cif.gz | 432.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8t3k.ent.gz pdb8t3k.ent.gz | 357.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8t3k.json.gz 8t3k.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/t3/8t3k https://data.pdbj.org/pub/pdb/validation_reports/t3/8t3k ftp://data.pdbj.org/pub/pdb/validation_reports/t3/8t3k ftp://data.pdbj.org/pub/pdb/validation_reports/t3/8t3k | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  41004MC  8fhkC  8fmvC  8fpfC  8szcC  8t1pC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 77369.898 Da / Num. of mol.: 1 / Mutation: C154A,C256A,C351A,E592Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Bacillus subtilis subsp. subtilis str. 168 (bacteria) Bacillus subtilis subsp. subtilis str. 168 (bacteria)Gene: yheH, BSU09720 / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli)References: UniProt: O07549, Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate | ||||
|---|---|---|---|---|---|
| #2: Protein | Mass: 67602.961 Da / Num. of mol.: 1 / Mutation: D500Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Bacillus subtilis subsp. subtilis str. 168 (bacteria) Bacillus subtilis subsp. subtilis str. 168 (bacteria)Gene: yheI, BSU09710 / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli)References: UniProt: O07550, Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate | ||||
| #3: Chemical | ChemComp-POV / (  POPC POPC#4: Chemical |  Adenosine triphosphate Adenosine triphosphateHas ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Heterodimeric BmrCD with ligands ATP and lipids / Type: COMPLEX / Entity ID: #1-#2 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:   Bacillus subtilis subsp. subtilis str. 168 (bacteria) Bacillus subtilis subsp. subtilis str. 168 (bacteria) |
| Source (recombinant) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 3 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2200 nm / Nominal defocus min: 400 nm / Cs Bright-field microscopy / Nominal defocus max: 2200 nm / Nominal defocus min: 400 nm / Cs : 2.7 mm : 2.7 mm |
| Specimen holder | Cryogen: NITROGEN |
| Image recording | Electron dose: 15 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
CTF correction | Type: NONE | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
3D reconstruction | Resolution: 3.33 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 76190 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller







 PDBj
PDBj





