[English] 日本語
 Yorodumi
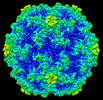
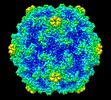
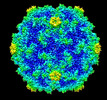
Yorodumi- PDB-8erk: Acheta domesticus segmented densovirus, high buoyancy (HB) capsid... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8erk | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acheta domesticus segmented densovirus, high buoyancy (HB) capsid, a mixed population of empty and immature full particles | ||||||||||||
 Components Components | Acheta domesticus segmented densovirus major capsid protein | ||||||||||||
 Keywords Keywords |  VIRUS / VIRUS /  parvovirus / parvovirus /  densovirus / virus capsid / densovirus / virus capsid /  virion / virion /  insect virus / insect virus /  capsid / capsid /  invertebrate invertebrate | ||||||||||||
| Biological species |  unclassified Densovirinae (virus) unclassified Densovirinae (virus) | ||||||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.5 Å cryo EM / Resolution: 2.5 Å | ||||||||||||
 Authors Authors | Penzes, J.J. / McKenna, R. / Tijssen, P. | ||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Bipartite genome and structural organization of the parvovirus Acheta domesticus segmented densovirus. Authors: Judit J Pénzes / Hanh T Pham / Paul Chipman / Emmanuel W Smith / Robert McKenna / Peter Tijssen /   Abstract: Parvoviruses (family Parvoviridae) are currently defined by a linear monopartite ssDNA genome, T = 1 icosahedral capsids, and distinct structural (VP) and non-structural (NS) protein expression ...Parvoviruses (family Parvoviridae) are currently defined by a linear monopartite ssDNA genome, T = 1 icosahedral capsids, and distinct structural (VP) and non-structural (NS) protein expression cassettes within their genome. We report the discovery of a parvovirus with a bipartite genome, Acheta domesticus segmented densovirus (AdSDV), isolated from house crickets (Acheta domesticus), in which it is pathogenic. We found that the AdSDV harbors its NS and VP cassettes on two separate genome segments. Its vp segment acquired a phospholipase A2-encoding gene, vpORF3, via inter-subfamily recombination, coding for a non-structural protein. We showed that the AdSDV evolved a highly complex transcription profile in response to its multipartite replication strategy compared to its monopartite ancestors. Our structural and molecular examinations revealed that the AdSDV packages one genome segment per particle. The cryo-EM structures of two empty- and one full-capsid population (3.3, 3.1 and 2.3 Å resolution) reveal a genome packaging mechanism, which involves an elongated C-terminal tail of the VP, "pinning" the ssDNA genome to the capsid interior at the twofold symmetry axis. This mechanism fundamentally differs from the capsid-DNA interactions previously seen in parvoviruses. This study provides new insights on the mechanism behind ssDNA genome segmentation and on the plasticity of parvovirus biology. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8erk.cif.gz 8erk.cif.gz | 118.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8erk.ent.gz pdb8erk.ent.gz | 92.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8erk.json.gz 8erk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/er/8erk https://data.pdbj.org/pub/pdb/validation_reports/er/8erk ftp://data.pdbj.org/pub/pdb/validation_reports/er/8erk ftp://data.pdbj.org/pub/pdb/validation_reports/er/8erk | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  28553MC  8er8C  8eu6C  8eu7C M: map data used to model this data C: citing same article ( |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 | x 60
|
| 2 |
|
| 3 | x 5
|
| 4 | x 6
|
| 5 | 
|
| Symmetry | Point symmetry: (Schoenflies symbol : I (icosahedral : I (icosahedral )) )) |
- Components
Components
| #1: Protein | Mass: 36291.906 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  unclassified Densovirinae (virus) unclassified Densovirinae (virus) |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: unclassified Densovirinae / Type: VIRUS / Entity ID: all / Source: NATURAL |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  unclassified Densovirinae (virus) unclassified Densovirinae (virus) |
| Details of virus | Empty: YES / Enveloped: NO / Isolate: SPECIES / Type: VIRION |
| Natural host | Organism: Acheta domesticus |
| Virus shell | Name: Acheta domesticus segmented densovirus capsid / Diameter: 210 nm / Triangulation number (T number): 1 |
| Buffer solution | pH: 7.4 |
| Buffer component | Conc.: 1 x / Name: Phosphate-buffered saline |
| Specimen | Conc.: 2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YESDetails: Deceased common house crickets, previously showing signs of paralysis, inability to jump, and inability to control movement |
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 |
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 90 % |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : OTHER / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM : OTHER / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: OTHER / Nominal defocus max: 4000 nm / Nominal defocus min: 650 nm / Cs : 2.7 mm : 2.7 mm |
| Image recording | Electron dose: 75 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 1127 |
| EM imaging optics | Phase plate: VOLTA PHASE PLATE |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.10_2155: / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
CTF correction | Type: PHASE FLIPPING ONLY | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 98920 | ||||||||||||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 2.5 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 59211 / Algorithm: FOURIER SPACE / Num. of class averages: 3 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 3UX1 Pdb chain-ID: A / Accession code: 3UX1 / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj

