[English] 日本語
 Yorodumi
Yorodumi- PDB-8d3h: Crystal structure of human Apoptosis-Inducing Factor (AIF) W196A ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8d3h | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human Apoptosis-Inducing Factor (AIF) W196A mutant complexed with 7-chloroquinolin-4-amine | ||||||||||||
 Components Components | (Apoptosis-inducing factor 1, ... ) x 2 ) x 2 | ||||||||||||
 Keywords Keywords |  OXIDOREDUCTASE / mitochondrial import / OXIDOREDUCTASE / mitochondrial import /  oxidative phosphorylation / oxidative phosphorylation /  SAXS SAXS | ||||||||||||
| Function / homology |  Function and homology information Function and homology information Oxidoreductases; Acting on NADH or NADPH; With unknown physiological acceptors / Oxidoreductases; Acting on NADH or NADPH; With unknown physiological acceptors /  regulation of apoptotic DNA fragmentation / mitochondrial respiratory chain complex assembly / protein import into mitochondrial intermembrane space / poly-ADP-D-ribose binding / NAD(P)H oxidase H2O2-forming activity / cellular response to aldosterone / positive regulation of necroptotic process / regulation of apoptotic DNA fragmentation / mitochondrial respiratory chain complex assembly / protein import into mitochondrial intermembrane space / poly-ADP-D-ribose binding / NAD(P)H oxidase H2O2-forming activity / cellular response to aldosterone / positive regulation of necroptotic process /  NADH dehydrogenase activity / oxidoreductase activity, acting on NAD(P)H ... NADH dehydrogenase activity / oxidoreductase activity, acting on NAD(P)H ... Oxidoreductases; Acting on NADH or NADPH; With unknown physiological acceptors / Oxidoreductases; Acting on NADH or NADPH; With unknown physiological acceptors /  regulation of apoptotic DNA fragmentation / mitochondrial respiratory chain complex assembly / protein import into mitochondrial intermembrane space / poly-ADP-D-ribose binding / NAD(P)H oxidase H2O2-forming activity / cellular response to aldosterone / positive regulation of necroptotic process / regulation of apoptotic DNA fragmentation / mitochondrial respiratory chain complex assembly / protein import into mitochondrial intermembrane space / poly-ADP-D-ribose binding / NAD(P)H oxidase H2O2-forming activity / cellular response to aldosterone / positive regulation of necroptotic process /  NADH dehydrogenase activity / oxidoreductase activity, acting on NAD(P)H / chromosome condensation / response to L-glutamate / mitochondrial respiratory chain complex I assembly / intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress / cellular response to nitric oxide / FAD binding / cellular response to estradiol stimulus / response to ischemia / NADH dehydrogenase activity / oxidoreductase activity, acting on NAD(P)H / chromosome condensation / response to L-glutamate / mitochondrial respiratory chain complex I assembly / intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress / cellular response to nitric oxide / FAD binding / cellular response to estradiol stimulus / response to ischemia /  mitochondrial intermembrane space / neuron differentiation / response to toxic substance / cellular response to hydrogen peroxide / activation of cysteine-type endopeptidase activity involved in apoptotic process / positive regulation of neuron apoptotic process / cellular response to hypoxia / neuron apoptotic process / mitochondrial intermembrane space / neuron differentiation / response to toxic substance / cellular response to hydrogen peroxide / activation of cysteine-type endopeptidase activity involved in apoptotic process / positive regulation of neuron apoptotic process / cellular response to hypoxia / neuron apoptotic process /  mitochondrial inner membrane / mitochondrial inner membrane /  protein dimerization activity / positive regulation of apoptotic process / apoptotic process / perinuclear region of cytoplasm / protein dimerization activity / positive regulation of apoptotic process / apoptotic process / perinuclear region of cytoplasm /  mitochondrion / mitochondrion /  DNA binding / DNA binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.51 Å MOLECULAR REPLACEMENT / Resolution: 2.51 Å | ||||||||||||
 Authors Authors | Brosey, C.A. / Tainer, J.A. | ||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Integrating early structural selection into chemical library screening for drug discovery with high-throughput small-angle X-ray scattering (SAXS) Authors: Brosey, C.A. / Link, T. / Shen, R. / Moiani, D. / Burnett, K. / Hura, G. / Jones, D.E. / Tainer, J.A. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8d3h.cif.gz 8d3h.cif.gz | 594 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8d3h.ent.gz pdb8d3h.ent.gz | 406.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8d3h.json.gz 8d3h.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/d3/8d3h https://data.pdbj.org/pub/pdb/validation_reports/d3/8d3h ftp://data.pdbj.org/pub/pdb/validation_reports/d3/8d3h ftp://data.pdbj.org/pub/pdb/validation_reports/d3/8d3h | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8d3eC  8d3gC  8d3iC  8d3jC  8d3kC  8d3nC  8d3oC  5kvhS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
|
- Components
Components
-Apoptosis-inducing factor 1, ... , 2 types, 2 molecules AB
| #1: Protein |  / Programmed cell death protein 8 / Programmed cell death protein 8Mass: 59371.434 Da / Num. of mol.: 1 / Mutation: W196A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: AIFM1, AIF, PDCD8 / Production host: Homo sapiens (human) / Gene: AIFM1, AIF, PDCD8 / Production host:   Escherichia coli BL21(DE3) (bacteria) / Variant (production host): Rosetta2 Escherichia coli BL21(DE3) (bacteria) / Variant (production host): Rosetta2References: UniProt: O95831,  Oxidoreductases; Acting on NADH or NADPH; With unknown physiological acceptors Oxidoreductases; Acting on NADH or NADPH; With unknown physiological acceptors |
|---|---|
| #2: Protein |  / Programmed cell death protein 8 / Programmed cell death protein 8Mass: 59387.434 Da / Num. of mol.: 1 / Mutation: W196A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: AIFM1, AIF, PDCD8 / Production host: Homo sapiens (human) / Gene: AIFM1, AIF, PDCD8 / Production host:   Escherichia coli BL21(DE3) (bacteria) / Variant (production host): Rosetta2 Escherichia coli BL21(DE3) (bacteria) / Variant (production host): Rosetta2References: UniProt: O95831,  Oxidoreductases; Acting on NADH or NADPH; With unknown physiological acceptors Oxidoreductases; Acting on NADH or NADPH; With unknown physiological acceptors |
-Non-polymers , 4 types, 185 molecules 
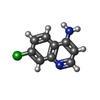





| #3: Chemical |  Flavin adenine dinucleotide Flavin adenine dinucleotide#4: Chemical | #5: Chemical | ChemComp-EDO / |  Ethylene glycol Ethylene glycol#6: Water | ChemComp-HOH / |  Water Water |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.58 Å3/Da / Density % sol: 52.3 % |
|---|---|
Crystal grow | Temperature: 295 K / Method: vapor diffusion, hanging drop / pH: 8.5 / Details: 0.1 M Tris, pH 8.5, 0.3 M Na2SO4, 18% PEG3350 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS-II NSLS-II  / Beamline: 17-ID-2 / Wavelength: 0.9793 Å / Beamline: 17-ID-2 / Wavelength: 0.9793 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Oct 1, 2021 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9793 Å / Relative weight: 1 : 0.9793 Å / Relative weight: 1 |
| Reflection | Resolution: 2.51→29.36 Å / Num. obs: 43738 / % possible obs: 99.6 % / Redundancy: 10.4 % / Biso Wilson estimate: 51.24 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.084 / Rpim(I) all: 0.027 / Rrim(I) all: 0.089 / Net I/σ(I): 18.7 |
| Reflection shell | Resolution: 2.51→2.61 Å / Rmerge(I) obs: 0.737 / Mean I/σ(I) obs: 3.1 / Num. unique obs: 4401 / CC1/2: 0.906 / Rpim(I) all: 0.235 / Rrim(I) all: 0.774 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5KVH Resolution: 2.51→29.36 Å / SU ML: 0.279 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 25.1996 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 62.67 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.51→29.36 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj





