Entry Database : PDB / ID : 7ztlTitle Crystal structure of a covalently linked Aurora-A N-Myc complex Aurora kinase A N-myc proto-oncogene protein Keywords / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 1.9 Å Authors Diebold, M. / Schindelin, H. Funding support Organization Grant number Country German Research Foundation (DFG) GRK2243
Journal : Acta Crystallogr D Struct Biol / Year : 2023Title : Crystal structure of a covalently linked Aurora-A-MYCN complex.Authors : Diebold, M. / Schonemann, L. / Eilers, M. / Sotriffer, C. / Schindelin, H. History Deposition May 11, 2022 Deposition site / Processing site Revision 1.0 Jan 18, 2023 Provider / Type Revision 1.1 Jan 31, 2024 Group / Refinement descriptionCategory / chem_comp_bond / pdbx_initial_refinement_modelRevision 1.2 Apr 3, 2024 Group / Category / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation_author.identifier_ORCID
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords TRANSFERASE / Crosslink / protein-protein-complex
TRANSFERASE / Crosslink / protein-protein-complex Function and homology information
Function and homology information regulation of centrosome cycle /
regulation of centrosome cycle /  axon hillock / spindle assembly involved in female meiosis I / cilium disassembly / positive regulation of oocyte maturation / spindle pole centrosome / histone H3S10 kinase activity ...regulation of inner ear auditory receptor cell differentiation / embryonic skeletal system morphogenesis / Interaction between PHLDA1 and AURKA /
axon hillock / spindle assembly involved in female meiosis I / cilium disassembly / positive regulation of oocyte maturation / spindle pole centrosome / histone H3S10 kinase activity ...regulation of inner ear auditory receptor cell differentiation / embryonic skeletal system morphogenesis / Interaction between PHLDA1 and AURKA /  regulation of centrosome cycle /
regulation of centrosome cycle /  axon hillock / spindle assembly involved in female meiosis I / cilium disassembly / positive regulation of oocyte maturation / spindle pole centrosome / histone H3S10 kinase activity / chromosome passenger complex /
axon hillock / spindle assembly involved in female meiosis I / cilium disassembly / positive regulation of oocyte maturation / spindle pole centrosome / histone H3S10 kinase activity / chromosome passenger complex /  pronucleus / astrocyte differentiation / positive regulation of programmed cell death /
pronucleus / astrocyte differentiation / positive regulation of programmed cell death /  meiotic spindle / mitotic centrosome separation /
meiotic spindle / mitotic centrosome separation /  germinal vesicle / protein localization to centrosome / anterior/posterior axis specification / centrosome localization / positive regulation of mesenchymal cell proliferation / neuron projection extension / spindle organization / embryonic digit morphogenesis / cartilage condensation / positive regulation of mitochondrial fission / Signaling by ALK / branching morphogenesis of an epithelial tube / mitotic spindle pole / SUMOylation of DNA replication proteins / negative regulation of astrocyte differentiation / negative regulation of reactive oxygen species metabolic process / spindle midzone / regulation of G2/M transition of mitotic cell cycle /
germinal vesicle / protein localization to centrosome / anterior/posterior axis specification / centrosome localization / positive regulation of mesenchymal cell proliferation / neuron projection extension / spindle organization / embryonic digit morphogenesis / cartilage condensation / positive regulation of mitochondrial fission / Signaling by ALK / branching morphogenesis of an epithelial tube / mitotic spindle pole / SUMOylation of DNA replication proteins / negative regulation of astrocyte differentiation / negative regulation of reactive oxygen species metabolic process / spindle midzone / regulation of G2/M transition of mitotic cell cycle /  centriole / protein serine/threonine/tyrosine kinase activity / positive regulation of mitotic cell cycle / positive regulation of mitotic nuclear division / AURKA Activation by TPX2 / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest / mitotic spindle organization / ciliary basal body /
centriole / protein serine/threonine/tyrosine kinase activity / positive regulation of mitotic cell cycle / positive regulation of mitotic nuclear division / AURKA Activation by TPX2 / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest / mitotic spindle organization / ciliary basal body /  regulation of cytokinesis / negative regulation of protein binding / regulation of signal transduction by p53 class mediator / epithelial cell proliferation / molecular function activator activity / positive regulation of epithelial cell proliferation /
regulation of cytokinesis / negative regulation of protein binding / regulation of signal transduction by p53 class mediator / epithelial cell proliferation / molecular function activator activity / positive regulation of epithelial cell proliferation /  liver regeneration / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / lung development / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 /
liver regeneration / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / lung development / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 /  regulation of protein stability / spindle microtubule /
regulation of protein stability / spindle microtubule /  mitotic spindle /
mitotic spindle /  kinetochore /
kinetochore /  kinase binding / positive regulation of miRNA transcription / response to wounding / spindle / G2/M transition of mitotic cell cycle / microtubule cytoskeleton /
kinase binding / positive regulation of miRNA transcription / response to wounding / spindle / G2/M transition of mitotic cell cycle / microtubule cytoskeleton /  Regulation of PLK1 Activity at G2/M Transition / sequence-specific double-stranded DNA binding / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / mitotic cell cycle / midbody / DNA-binding transcription activator activity, RNA polymerase II-specific / basolateral plasma membrane / peptidyl-serine phosphorylation / proteasome-mediated ubiquitin-dependent protein catabolic process / Regulation of TP53 Activity through Phosphorylation / protein autophosphorylation /
Regulation of PLK1 Activity at G2/M Transition / sequence-specific double-stranded DNA binding / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / mitotic cell cycle / midbody / DNA-binding transcription activator activity, RNA polymerase II-specific / basolateral plasma membrane / peptidyl-serine phosphorylation / proteasome-mediated ubiquitin-dependent protein catabolic process / Regulation of TP53 Activity through Phosphorylation / protein autophosphorylation /  postsynaptic density /
postsynaptic density /  protein dimerization activity /
protein dimerization activity /  non-specific serine/threonine protein kinase / DNA-binding transcription factor activity, RNA polymerase II-specific /
non-specific serine/threonine protein kinase / DNA-binding transcription factor activity, RNA polymerase II-specific /  protein kinase activity / RNA polymerase II cis-regulatory region sequence-specific DNA binding / protein heterodimerization activity / DNA-binding transcription factor activity /
protein kinase activity / RNA polymerase II cis-regulatory region sequence-specific DNA binding / protein heterodimerization activity / DNA-binding transcription factor activity /  cell division /
cell division /  protein phosphorylation / negative regulation of gene expression / protein serine kinase activity / protein serine/threonine kinase activity /
protein phosphorylation / negative regulation of gene expression / protein serine kinase activity / protein serine/threonine kinase activity /  centrosome / glutamatergic synapse / apoptotic process /
centrosome / glutamatergic synapse / apoptotic process /  ubiquitin protein ligase binding /
ubiquitin protein ligase binding /  chromatin / positive regulation of gene expression /
chromatin / positive regulation of gene expression /  nucleolus / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process /
nucleolus / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process /  protein kinase binding / perinuclear region of cytoplasm / positive regulation of DNA-templated transcription / positive regulation of transcription by RNA polymerase II /
protein kinase binding / perinuclear region of cytoplasm / positive regulation of DNA-templated transcription / positive regulation of transcription by RNA polymerase II /  DNA binding
DNA binding
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å
MOLECULAR REPLACEMENT / Resolution: 1.9 Å  Authors
Authors Germany, 1items
Germany, 1items  Citation
Citation Journal: Acta Crystallogr D Struct Biol / Year: 2023
Journal: Acta Crystallogr D Struct Biol / Year: 2023 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7ztl.cif.gz
7ztl.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7ztl.ent.gz
pdb7ztl.ent.gz PDB format
PDB format 7ztl.json.gz
7ztl.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/zt/7ztl
https://data.pdbj.org/pub/pdb/validation_reports/zt/7ztl ftp://data.pdbj.org/pub/pdb/validation_reports/zt/7ztl
ftp://data.pdbj.org/pub/pdb/validation_reports/zt/7ztl
 F&H Search
F&H Search Links
Links Assembly
Assembly
 Components
Components / Aurora 2 / Aurora/IPL1-related kinase 1 / ARK-1 / Aurora-related kinase 1 / hARK1 / Breast tumor- ...Aurora 2 / Aurora/IPL1-related kinase 1 / ARK-1 / Aurora-related kinase 1 / hARK1 / Breast tumor-amplified kinase / Serine/threonine-protein kinase 15 / Serine/threonine-protein kinase 6 / Serine/threonine-protein kinase aurora-A
/ Aurora 2 / Aurora/IPL1-related kinase 1 / ARK-1 / Aurora-related kinase 1 / hARK1 / Breast tumor- ...Aurora 2 / Aurora/IPL1-related kinase 1 / ARK-1 / Aurora-related kinase 1 / hARK1 / Breast tumor-amplified kinase / Serine/threonine-protein kinase 15 / Serine/threonine-protein kinase 6 / Serine/threonine-protein kinase aurora-A
 Homo sapiens (human)
Homo sapiens (human)
 Escherichia coli (E. coli)
Escherichia coli (E. coli) non-specific serine/threonine protein kinase
non-specific serine/threonine protein kinase
 Homo sapiens (human) / References: UniProt: P04198
Homo sapiens (human) / References: UniProt: P04198


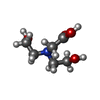









 Adenosine diphosphate
Adenosine diphosphate Glycerol
Glycerol Bicine
Bicine Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  PETRA III, EMBL c/o DESY
PETRA III, EMBL c/o DESY  / Beamline: P13 (MX1) / Wavelength: 0.9762 Å
/ Beamline: P13 (MX1) / Wavelength: 0.9762 Å : 0.9762 Å / Relative weight: 1
: 0.9762 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller



 PDBj
PDBj




















