+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7wwu | ||||||
|---|---|---|---|---|---|---|---|
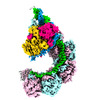
| Title | ICP1 Csy complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA BINDING PROTEIN/RNA /  CRISPR-Cas system / CRISPR-Cas system /  RNA / RNA /  RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | ||||||
 Authors Authors | Zhang, M. / Peng, R. | ||||||
| Funding support |  China, 1items China, 1items
| ||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage. Authors: Manling Zhang / Ruchao Peng / Qi Peng / Sheng Liu / Zhiteng Li / Yuqin Zhang / Hao Song / Jia Yang / Xiao Xing / Peiyi Wang / Jianxun Qi / George F Gao /  Abstract: CRISPR-Cas systems are widespread adaptive antiviral systems used in prokaryotes. Some phages, in turn, although have small genomes can economize the use of genetic space to encode compact or ...CRISPR-Cas systems are widespread adaptive antiviral systems used in prokaryotes. Some phages, in turn, although have small genomes can economize the use of genetic space to encode compact or incomplete CRISPR-Cas systems to inhibit the host and establish infection. Phage ICP1, infecting , encodes a compact type I-F CRISPR-Cas system to suppress the antiphage mobile genetic element in the host genome. However, the mechanism by which this compact system recognizes the target DNA and executes interference remains elusive. Here, we present the electron cryo-microscopy (cryo-EM) structures of both apo- and DNA-bound ICP1 surveillance complexes (Aka Csy complex). Unlike most other type I surveillance complexes, the ICP1 Csy complex lacks the Cas11 subunit or a structurally homologous domain, which is crucial for dsDNA binding and Cas3 activation in other type I CRISPR-Cas systems. Structural and functional analyses revealed that the compact ICP1 Csy complex alone is inefficient in binding to dsDNA targets, presumably stalled at a partial R-loop conformation. The presence of Cas2/3 facilitates dsDNA binding and allows effective dsDNA target cleavage. Additionally, we found that Cas2/3 efficiently cleaved the dsDNA target presented by the ICP1 Csy complex, but not vice versa. These findings suggest a unique mechanism for target dsDNA binding and cleavage by the compact phage-derived CRISPR-Cas system. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7wwu.cif.gz 7wwu.cif.gz | 455.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7wwu.ent.gz pdb7wwu.ent.gz | 370.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7wwu.json.gz 7wwu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ww/7wwu https://data.pdbj.org/pub/pdb/validation_reports/ww/7wwu ftp://data.pdbj.org/pub/pdb/validation_reports/ww/7wwu ftp://data.pdbj.org/pub/pdb/validation_reports/ww/7wwu | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  32873MC  7wkoC  7wkpC  7wwvC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 22841.510 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Vibrio phage ICP1_2011_A (virus) / Gene: csy1, ICP12011A_088 Vibrio phage ICP1_2011_A (virus) / Gene: csy1, ICP12011A_088Production host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: M1R2X3 | ||||
|---|---|---|---|---|---|
| #2: Protein | Mass: 29833.129 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Vibrio phage ICP1_2011_A (virus) / Gene: csy2, ICP12011A_087 Vibrio phage ICP1_2011_A (virus) / Gene: csy2, ICP12011A_087Production host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: M1QWL5 | ||||
| #3: Protein | Mass: 35830.867 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Vibrio phage ICP1_2011_A (virus) / Gene: csy3, ICP12011A_086 Vibrio phage ICP1_2011_A (virus) / Gene: csy3, ICP12011A_086Production host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: M1Q7R8 #4: Protein | | Mass: 21168.271 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Vibrio phage ICP1_2011_A (virus) / Gene: csy4, ICP12011A_085 Vibrio phage ICP1_2011_A (virus) / Gene: csy4, ICP12011A_085Production host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: M1R9H3 #5: RNA chain | | Mass: 19351.408 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus)Production host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: GenBank: 452118997 |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: apo-ICP1 Csy complex / Type: COMPLEX / Entity ID: all / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) |
| Source (recombinant) | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 1800 nm Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 1800 nm |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.19.2_4158: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
3D reconstruction | Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 463686 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj





























