+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7t3k | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Csy-AcrIF24 dimer | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE/RNA / Csy / Cascade /  CRISPR / CRISPR /  anti-CRISPR / AcrIF24 / HYDROLASE-RNA complex anti-CRISPR / AcrIF24 / HYDROLASE-RNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements /  endonuclease activity / defense response to virus / endonuclease activity / defense response to virus /  Hydrolases; Acting on ester bonds / Hydrolases; Acting on ester bonds /  RNA binding RNA bindingSimilarity search - Function | ||||||
| Biological species |   Pseudomonas (bacteria) Pseudomonas (bacteria) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | ||||||
 Authors Authors | Mukherjee, I.A. / Chang, L. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2022 Journal: Nat Chem Biol / Year: 2022Title: Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor. Authors: Indranil Arun Mukherjee / Clinton Gabel / Nicholas Noinaj / Joseph Bondy-Denomy / Leifu Chang /  Abstract: Anti-CRISPR (Acr) proteins are encoded by phages to inactivate CRISPR-Cas systems of bacteria and archaea and are used to enhance the CRISPR toolbox for genome editing. Here we report the structure ...Anti-CRISPR (Acr) proteins are encoded by phages to inactivate CRISPR-Cas systems of bacteria and archaea and are used to enhance the CRISPR toolbox for genome editing. Here we report the structure and mechanism of AcrIF24, an Acr protein that inhibits the type I-F CRISPR-Cas system from Pseudomonas aeruginosa. AcrIF24 is a homodimer that associates with two copies of the surveillance complex (Csy) and prevents the hybridization between CRISPR RNA and target DNA. Furthermore, AcrIF24 functions as an anti-CRISPR-associated (Aca) protein to repress the transcription of the acrIF23-acrIF24 operon. Alone or in complex with Csy, AcrIF24 is capable of binding to the acrIF23-acrIF24 promoter DNA with nanomolar affinity. The structure of a Csy-AcrIF24-promoter DNA complex at 2.7 Å reveals the mechanism for transcriptional suppression. Our results reveal that AcrIF24 functions as an Acr-Aca fusion protein, and they extend understanding of the diverse mechanisms used by Acr proteins. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7t3k.cif.gz 7t3k.cif.gz | 1.2 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7t3k.ent.gz pdb7t3k.ent.gz | 1008.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7t3k.json.gz 7t3k.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/t3/7t3k https://data.pdbj.org/pub/pdb/validation_reports/t3/7t3k ftp://data.pdbj.org/pub/pdb/validation_reports/t3/7t3k ftp://data.pdbj.org/pub/pdb/validation_reports/t3/7t3k | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 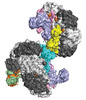 25661MC  7t3jC  7t3lC  7tawC  7taxC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-CRISPR-associated ... , 2 types, 4 molecules AaCc
| #1: Protein | Mass: 49194.168 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas (bacteria) / Gene: csy1, PA14_33330 / Production host: Pseudomonas (bacteria) / Gene: csy1, PA14_33330 / Production host:   Escherichia (bacteria) / References: UniProt: Q02ML9 Escherichia (bacteria) / References: UniProt: Q02ML9#3: Protein | Mass: 21427.504 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas (bacteria) / Gene: cas6f, csy4, PA14_33300 / Production host: Pseudomonas (bacteria) / Gene: cas6f, csy4, PA14_33300 / Production host:   Escherichia (bacteria) Escherichia (bacteria)References: UniProt: Q02MM2,  Hydrolases; Acting on ester bonds Hydrolases; Acting on ester bonds |
|---|
-CRISPR type I-F/YPEST-associated protein ... , 2 types, 14 molecules BbDEFGHIdefghi
| #2: Protein | Mass: 36244.074 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas (bacteria) Pseudomonas (bacteria)Gene: csy2, ALP65_00953, IPC1598_32520, IPC29_26970, PA52Ts2_3638, PACL_0128 Production host:   Escherichia (bacteria) / References: UniProt: B3G161 Escherichia (bacteria) / References: UniProt: B3G161#4: Protein | Mass: 39778.594 Da / Num. of mol.: 12 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas (bacteria) / Gene: PA52Ts2_3639 / Production host: Pseudomonas (bacteria) / Gene: PA52Ts2_3639 / Production host:   Escherichia (bacteria) / References: UniProt: A0A444M080 Escherichia (bacteria) / References: UniProt: A0A444M080 |
|---|
-Protein / RNA chain , 2 types, 4 molecules JKMm
| #5: Protein | Mass: 24995.271 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas (bacteria) / Production host: Pseudomonas (bacteria) / Production host:   Escherichia (bacteria) Escherichia (bacteria)#6: RNA chain | Mass: 19538.586 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas (bacteria) / Production host: Pseudomonas (bacteria) / Production host:   Escherichia (bacteria) Escherichia (bacteria) |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Csy-AcrIF / Type: COMPLEX / Entity ID: all / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 400 kDa/nm / Experimental value: NO |
| Source (natural) | Organism:   Pseudomonas (bacteria) Pseudomonas (bacteria) |
| Source (recombinant) | Organism:   Escherichia (bacteria) Escherichia (bacteria) |
| Buffer solution | pH: 7.5 |
| Specimen | Conc.: 3.5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: OTHER / Nominal defocus max: 2000 nm / Nominal defocus min: 800 nm |
| Image recording | Electron dose: 54 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.20_4459: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||
CTF correction | Type: NONE | ||||||||||||||||||||||||
3D reconstruction | Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 58133 / Symmetry type: POINT | ||||||||||||||||||||||||
| Atomic model building | Space: REAL | ||||||||||||||||||||||||
| Atomic model building | PDB-ID: 7JZW | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller







 PDBj
PDBj






























