[English] 日本語
 Yorodumi
Yorodumi- PDB-7p55: NMR structure of human ACE2 21-42 fragment in HFIP/water 50/50 v/v -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7p55 | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR structure of human ACE2 21-42 fragment in HFIP/water 50/50 v/v | ||||||
 Components Components | Processed angiotensin-converting enzyme 2 | ||||||
 Keywords Keywords |  ANTIVIRAL PROTEIN / ANTIVIRAL PROTEIN /  STRUCTURE FROM CYANA 2.1 / STRUCTURE FROM CYANA 2.1 /  angiotensin converting enzyme 2 / angiotensin converting enzyme 2 /  ACE2 / ACE2 /  SARS-CoV-2 / SARS-CoV-2 /  COVID-19 COVID-19 | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of amino acid transport /  angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane /  Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / positive regulation of gap junction assembly / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / positive regulation of gap junction assembly /  regulation of vasoconstriction / regulation of cardiac conduction ...positive regulation of amino acid transport / regulation of vasoconstriction / regulation of cardiac conduction ...positive regulation of amino acid transport /  angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane /  Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / positive regulation of gap junction assembly / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / positive regulation of gap junction assembly /  regulation of vasoconstriction / regulation of cardiac conduction / peptidyl-dipeptidase activity / angiotensin maturation / maternal process involved in female pregnancy / regulation of vasoconstriction / regulation of cardiac conduction / peptidyl-dipeptidase activity / angiotensin maturation / maternal process involved in female pregnancy /  metallocarboxypeptidase activity / Metabolism of Angiotensinogen to Angiotensins / Attachment and Entry / metallocarboxypeptidase activity / Metabolism of Angiotensinogen to Angiotensins / Attachment and Entry /  carboxypeptidase activity / negative regulation of signaling receptor activity / positive regulation of cardiac muscle contraction / regulation of cytokine production / carboxypeptidase activity / negative regulation of signaling receptor activity / positive regulation of cardiac muscle contraction / regulation of cytokine production /  viral life cycle / blood vessel diameter maintenance / brush border membrane / regulation of transmembrane transporter activity / negative regulation of smooth muscle cell proliferation / viral life cycle / blood vessel diameter maintenance / brush border membrane / regulation of transmembrane transporter activity / negative regulation of smooth muscle cell proliferation /  cilium / negative regulation of ERK1 and ERK2 cascade / endocytic vesicle membrane / cilium / negative regulation of ERK1 and ERK2 cascade / endocytic vesicle membrane /  metallopeptidase activity / positive regulation of reactive oxygen species metabolic process / virus receptor activity / regulation of cell population proliferation / metallopeptidase activity / positive regulation of reactive oxygen species metabolic process / virus receptor activity / regulation of cell population proliferation /  regulation of inflammatory response / regulation of inflammatory response /  endopeptidase activity / Induction of Cell-Cell Fusion / Potential therapeutics for SARS / entry receptor-mediated virion attachment to host cell / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / endopeptidase activity / Induction of Cell-Cell Fusion / Potential therapeutics for SARS / entry receptor-mediated virion attachment to host cell / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / receptor-mediated virion attachment to host cell / symbiont entry into host cell / membrane fusion / receptor-mediated virion attachment to host cell / symbiont entry into host cell /  membrane raft / apical plasma membrane / membrane raft / apical plasma membrane /  endoplasmic reticulum lumen / endoplasmic reticulum lumen /  cell surface / cell surface /  extracellular space / extracellular exosome / zinc ion binding / extracellular region / extracellular space / extracellular exosome / zinc ion binding / extracellular region /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION NMR / torsion angle dynamics SOLUTION NMR / torsion angle dynamics | ||||||
 Authors Authors | Santoro, A. / Buonocore, M. / Grimaldi, M. / D'Ursi, A.M. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: Heliyon / Year: 2022 Journal: Heliyon / Year: 2022Title: Structural analysis of a simplified model reproducing SARS-CoV-2 S RBD/ACE2 binding site. Authors: Buonocore, M. / Santoro, A. / Grimaldi, M. / Covelli, V. / Firoznezhad, M. / Rodriquez, M. / Santin, M. / D'Ursi, A.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7p55.cif.gz 7p55.cif.gz | 363.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7p55.ent.gz pdb7p55.ent.gz | 274.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7p55.json.gz 7p55.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/p5/7p55 https://data.pdbj.org/pub/pdb/validation_reports/p5/7p55 ftp://data.pdbj.org/pub/pdb/validation_reports/p5/7p55 ftp://data.pdbj.org/pub/pdb/validation_reports/p5/7p55 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7p5gC  7p5qC  7p5sC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 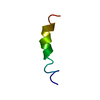
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 2718.941 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Homo sapiens (human) / References: UniProt: Q9BYF1 Homo sapiens (human) / References: UniProt: Q9BYF1 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Type: solution Contents: 2.5 mM ACE(21-42), 40 % v/v H2O, 10 % v/v [U-100% 2H] D2O, 50 v/v Hexafluoroisopropanol, 50% hexafluoroisopropanol/40% H2O/10% D2O Label: ACE(21-42) / Solvent system: 50% hexafluoroisopropanol/40% H2O/10% D2O | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||
| Sample conditions | Ionic strength: 0 Not defined / Label: condition_1 / pH: 2.5 / PH err: 0.1 / Pressure: 1 atm / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE II / Manufacturer: Bruker / Model : AVANCE II / Field strength: 500 MHz : AVANCE II / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics / Software ordinal: 3 | |||||||||||||||
| NMR representative | Selection criteria: target function | |||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 50 / Conformers submitted total number: 50 |
 Movie
Movie Controller
Controller


 PDBj
PDBj


