[English] 日本語
 Yorodumi
Yorodumi- PDB-6y2h: The crystal structure of human chloride intracellular channel pro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6y2h | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
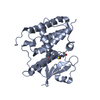
| Title | The crystal structure of human chloride intracellular channel protein 5 | |||||||||||||||
 Components Components | Chloride intracellular channel protein 5 Chloride channel Chloride channel | |||||||||||||||
 Keywords Keywords |  TRANSPORT PROTEIN / TRANSPORT PROTEIN /  Chloride intracellular channel / Chloride intracellular channel /  CLIC / CLIC /  metamorphic protein metamorphic protein | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationstereocilium bundle / Sensory processing of sound by outer hair cells of the cochlea / Sensory processing of sound by inner hair cells of the cochlea / chloride transport / response to stimulus /  chloride channel activity / chloride channel activity /  chloride channel complex / chloride channel complex /  visual perception / female pregnancy / sensory perception of sound ...stereocilium bundle / Sensory processing of sound by outer hair cells of the cochlea / Sensory processing of sound by inner hair cells of the cochlea / chloride transport / response to stimulus / visual perception / female pregnancy / sensory perception of sound ...stereocilium bundle / Sensory processing of sound by outer hair cells of the cochlea / Sensory processing of sound by inner hair cells of the cochlea / chloride transport / response to stimulus /  chloride channel activity / chloride channel activity /  chloride channel complex / chloride channel complex /  visual perception / female pregnancy / sensory perception of sound / visual perception / female pregnancy / sensory perception of sound /  actin cytoskeleton / actin cytoskeleton /  cell cortex / apical plasma membrane / cell cortex / apical plasma membrane /  centrosome / centrosome /  Golgi apparatus / Golgi apparatus /  mitochondrion / extracellular exosome mitochondrion / extracellular exosomeSimilarity search - Function | |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.154 Å MOLECULAR REPLACEMENT / Resolution: 2.154 Å | |||||||||||||||
 Authors Authors | Ferofontov, A. / Giladi, M. / Haitin, Y. | |||||||||||||||
| Funding support |  Israel, 4items Israel, 4items
| |||||||||||||||
 Citation Citation |  Journal: Faseb J. / Year: 2020 Journal: Faseb J. / Year: 2020Title: Conserved cysteine dioxidation enhances membrane interaction of human Cl - intracellular channel 5. Authors: Ferofontov, A. / Vankova, P. / Man, P. / Giladi, M. / Haitin, Y. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6y2h.cif.gz 6y2h.cif.gz | 355.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6y2h.ent.gz pdb6y2h.ent.gz | 288.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6y2h.json.gz 6y2h.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/y2/6y2h https://data.pdbj.org/pub/pdb/validation_reports/y2/6y2h ftp://data.pdbj.org/pub/pdb/validation_reports/y2/6y2h ftp://data.pdbj.org/pub/pdb/validation_reports/y2/6y2h | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2aheS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Ens-ID: 1
|
 Movie
Movie Controller
Controller











 PDBj
PDBj



