+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6wml | ||||||
|---|---|---|---|---|---|---|---|
| Title | Human TLR8 bound to the potent agonist, GS-9688 (Selgantolimod) | ||||||
 Components Components | Toll-like receptor 8 | ||||||
 Keywords Keywords |  IMMUNE SYSTEM / Selgantolimod / IMMUNE SYSTEM / Selgantolimod /  Toll-like Receptor / Immune Activator / Receptor agonist Toll-like Receptor / Immune Activator / Receptor agonist | ||||||
| Function / homology |  Function and homology information Function and homology informationToll Like Receptor 7/8 (TLR7/8) Cascade / toll-like receptor 8 signaling pathway / negative regulation of interleukin-12 production / endolysosome membrane / positive regulation of innate immune response / Trafficking and processing of endosomal TLR /  pattern recognition receptor activity / pattern recognition receptor activity /  toll-like receptor signaling pathway / positive regulation of interferon-alpha production / immunoglobulin mediated immune response ...Toll Like Receptor 7/8 (TLR7/8) Cascade / toll-like receptor 8 signaling pathway / negative regulation of interleukin-12 production / endolysosome membrane / positive regulation of innate immune response / Trafficking and processing of endosomal TLR / toll-like receptor signaling pathway / positive regulation of interferon-alpha production / immunoglobulin mediated immune response ...Toll Like Receptor 7/8 (TLR7/8) Cascade / toll-like receptor 8 signaling pathway / negative regulation of interleukin-12 production / endolysosome membrane / positive regulation of innate immune response / Trafficking and processing of endosomal TLR /  pattern recognition receptor activity / pattern recognition receptor activity /  toll-like receptor signaling pathway / positive regulation of interferon-alpha production / immunoglobulin mediated immune response / canonical NF-kappaB signal transduction / positive regulation of interferon-beta production / positive regulation of interleukin-1 beta production / positive regulation of interleukin-8 production / toll-like receptor signaling pathway / positive regulation of interferon-alpha production / immunoglobulin mediated immune response / canonical NF-kappaB signal transduction / positive regulation of interferon-beta production / positive regulation of interleukin-1 beta production / positive regulation of interleukin-8 production /  regulation of protein phosphorylation / response to virus / positive regulation of interleukin-6 production / cellular response to mechanical stimulus / positive regulation of type II interferon production / regulation of protein phosphorylation / response to virus / positive regulation of interleukin-6 production / cellular response to mechanical stimulus / positive regulation of type II interferon production /  double-stranded RNA binding / double-stranded RNA binding /  signaling receptor activity / defense response to virus / signaling receptor activity / defense response to virus /  single-stranded RNA binding / endosome membrane / single-stranded RNA binding / endosome membrane /  inflammatory response / external side of plasma membrane / inflammatory response / external side of plasma membrane /  Golgi membrane / Golgi membrane /  innate immune response / endoplasmic reticulum membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / innate immune response / endoplasmic reticulum membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses /  DNA binding / DNA binding /  RNA binding / identical protein binding / RNA binding / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.5 Å MOLECULAR REPLACEMENT / Resolution: 2.5 Å | ||||||
 Authors Authors | Appleby, T.C. / Perry, J.K. / Mish, M. / Villasenor, A.G. / Mackman, R.L. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2020 Journal: J.Med.Chem. / Year: 2020Title: Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B. Authors: Mackman, R.L. / Mish, M. / Chin, G. / Perry, J.K. / Appleby, T. / Aktoudianakis, V. / Metobo, S. / Pyun, P. / Niu, C. / Daffis, S. / Yu, H. / Zheng, J. / Villasenor, A.G. / Zablocki, J. / ...Authors: Mackman, R.L. / Mish, M. / Chin, G. / Perry, J.K. / Appleby, T. / Aktoudianakis, V. / Metobo, S. / Pyun, P. / Niu, C. / Daffis, S. / Yu, H. / Zheng, J. / Villasenor, A.G. / Zablocki, J. / Chamberlain, J. / Jin, H. / Lee, G. / Suekawa-Pirrone, K. / Santos, R. / Delaney 4th, W.E. / Fletcher, S.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6wml.cif.gz 6wml.cif.gz | 737.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6wml.ent.gz pdb6wml.ent.gz | 485.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6wml.json.gz 6wml.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wm/6wml https://data.pdbj.org/pub/pdb/validation_reports/wm/6wml ftp://data.pdbj.org/pub/pdb/validation_reports/wm/6wml ftp://data.pdbj.org/pub/pdb/validation_reports/wm/6wml | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3w3kS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein |  Mass: 92835.367 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: TLR8, UNQ249/PRO286 / Production host: Homo sapiens (human) / Gene: TLR8, UNQ249/PRO286 / Production host:   Drosophila melanogaster (fruit fly) / References: UniProt: Q9NR97 Drosophila melanogaster (fruit fly) / References: UniProt: Q9NR97 |
|---|
-Sugars , 7 types, 38 molecules 


| #2: Polysaccharide | alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1- ...alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose / Mass: 748.682 Da / Num. of mol.: 1 / Mass: 748.682 Da / Num. of mol.: 1Source method: isolated from a genetically manipulated source | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #3: Polysaccharide | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose / Mass: 383.349 Da / Num. of mol.: 1 / Mass: 383.349 Da / Num. of mol.: 1Source method: isolated from a genetically manipulated source | ||||||||
| #4: Polysaccharide | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta- ...beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose  / Mass: 586.542 Da / Num. of mol.: 7 / Mass: 586.542 Da / Num. of mol.: 7Source method: isolated from a genetically manipulated source #5: Polysaccharide |  / Mass: 748.682 Da / Num. of mol.: 2 / Mass: 748.682 Da / Num. of mol.: 2Source method: isolated from a genetically manipulated source #6: Polysaccharide | alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2- ...alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose |  / Mass: 910.823 Da / Num. of mol.: 1 / Mass: 910.823 Da / Num. of mol.: 1Source method: isolated from a genetically manipulated source #7: Sugar | ChemComp-MAN /  Mannose Mannose#8: Sugar | ChemComp-NAG /  N-Acetylglucosamine N-Acetylglucosamine |
-Non-polymers , 2 types, 428 molecules 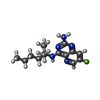


| #9: Chemical | ChemComp-U57 / ( #10: Water | ChemComp-HOH / |  Water Water |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.84 Å3/Da / Density % sol: 56.62 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 5 Details: 4% (w/v) PEG 3350 0.2 M potassium formate 0.1 M sodium citrate, pH 5.0 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.1 / Wavelength: 0.9762 Å / Beamline: 5.0.1 / Wavelength: 0.9762 Å |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Nov 10, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9762 Å / Relative weight: 1 : 0.9762 Å / Relative weight: 1 |
| Reflection | Resolution: 2→47.86 Å / Num. obs: 270193 / % possible obs: 98.1 % / Redundancy: 3.2 % / Biso Wilson estimate: 48.39 Å2 / CC1/2: 0.99 / Rrim(I) all: 0.06 / Net I/σ(I): 17.44 |
| Reflection shell | Resolution: 2.5→2.65 Å / Num. unique obs: 22744 / CC1/2: 0.84 / Rrim(I) all: 0.498 / % possible all: 98.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3W3K Resolution: 2.5→47.86 Å / SU ML: 0.3222 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 26.9066 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 57.09 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→47.86 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj











