| Entry | Database: PDB / ID: 6wax
|
|---|
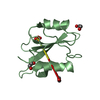
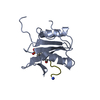
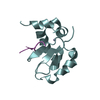
| Title | C-terminal SH2 domain of p120RasGAP |
|---|
 Components Components | Ras GTPase-activating protein 1 |
|---|
 Keywords Keywords |  SIGNALING PROTEIN / SIGNALING PROTEIN /  SH2 domain / SH2 domain /  RasGAP / RasGAP /  phosphopeptide / phosphopeptide /  phosphotyrosine phosphotyrosine |
|---|
| Function / homology |  Function and homology information Function and homology information
regulation of RNA metabolic process / regulation of actin filament polymerization / potassium channel inhibitor activity / negative regulation of cell adhesion / blood vessel morphogenesis / negative regulation of cell-matrix adhesion / mitotic cytokinesis / ephrin receptor signaling pathway /  vasculogenesis / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases ...regulation of RNA metabolic process / regulation of actin filament polymerization / potassium channel inhibitor activity / negative regulation of cell adhesion / blood vessel morphogenesis / negative regulation of cell-matrix adhesion / mitotic cytokinesis / ephrin receptor signaling pathway / vasculogenesis / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases ...regulation of RNA metabolic process / regulation of actin filament polymerization / potassium channel inhibitor activity / negative regulation of cell adhesion / blood vessel morphogenesis / negative regulation of cell-matrix adhesion / mitotic cytokinesis / ephrin receptor signaling pathway /  vasculogenesis / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / ruffle / EPHB-mediated forward signaling / phosphotyrosine residue binding / Downstream signal transduction / vasculogenesis / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / ruffle / EPHB-mediated forward signaling / phosphotyrosine residue binding / Downstream signal transduction /  GTPase activator activity / VEGFR2 mediated cell proliferation / Regulation of RAS by GAPs / GTPase activator activity / VEGFR2 mediated cell proliferation / Regulation of RAS by GAPs /  GTPase binding / regulation of cell shape / negative regulation of neuron apoptotic process / intracellular signal transduction / GTPase binding / regulation of cell shape / negative regulation of neuron apoptotic process / intracellular signal transduction /  signaling receptor binding / signaling receptor binding /  GTPase activity / negative regulation of apoptotic process / GTPase activity / negative regulation of apoptotic process /  signal transduction / signal transduction /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasmSimilarity search - Function cytoplasmSimilarity search - Function Ras GTPase-activating protein 1, N-terminal SH2 domain /  RasGAP, SH3 domain / Ras GTPase-activating protein 1, C-terminal SH2 domain / Ras GTPase-activating protein / Ras GTPase-activating protein, conserved site / Ras GTPase-activating proteins domain signature. / RasGAP, SH3 domain / Ras GTPase-activating protein 1, C-terminal SH2 domain / Ras GTPase-activating protein / Ras GTPase-activating protein, conserved site / Ras GTPase-activating proteins domain signature. /  GTPase-activator protein for Ras-like GTPase / Ras GTPase-activating proteins profile. / GTPase-activator protein for Ras-like GTPases / Ras GTPase-activating domain ...Ras GTPase-activating protein 1, N-terminal SH2 domain / GTPase-activator protein for Ras-like GTPase / Ras GTPase-activating proteins profile. / GTPase-activator protein for Ras-like GTPases / Ras GTPase-activating domain ...Ras GTPase-activating protein 1, N-terminal SH2 domain /  RasGAP, SH3 domain / Ras GTPase-activating protein 1, C-terminal SH2 domain / Ras GTPase-activating protein / Ras GTPase-activating protein, conserved site / Ras GTPase-activating proteins domain signature. / RasGAP, SH3 domain / Ras GTPase-activating protein 1, C-terminal SH2 domain / Ras GTPase-activating protein / Ras GTPase-activating protein, conserved site / Ras GTPase-activating proteins domain signature. /  GTPase-activator protein for Ras-like GTPase / Ras GTPase-activating proteins profile. / GTPase-activator protein for Ras-like GTPases / Ras GTPase-activating domain / Rho GTPase activation protein / GTPase-activator protein for Ras-like GTPase / Ras GTPase-activating proteins profile. / GTPase-activator protein for Ras-like GTPases / Ras GTPase-activating domain / Rho GTPase activation protein /  SH2 domain / SHC Adaptor Protein / SH2 domain / SHC Adaptor Protein /  C2 domain / Protein kinase C conserved region 2 (CalB) / C2 domain / Protein kinase C conserved region 2 (CalB) /  C2 domain / C2 domain profile. / C2 domain / C2 domain profile. /  PH domain / C2 domain superfamily / PH domain profile. / PH domain / C2 domain superfamily / PH domain profile. /  Pleckstrin homology domain. / Pleckstrin homology domain. /  Pleckstrin homology domain / Pleckstrin homology domain /  SH3 domain / SH3 domain /  SH2 domain / Src homology 2 (SH2) domain profile. / Src homology 2 domains / SH2 domain / Src homology 2 (SH2) domain profile. / Src homology 2 domains /  SH2 domain / Src homology 3 domains / SH2 domain superfamily / SH3-like domain superfamily / Src homology 3 (SH3) domain profile. / SH2 domain / Src homology 3 domains / SH2 domain superfamily / SH3-like domain superfamily / Src homology 3 (SH3) domain profile. /  SH3 domain / PH-like domain superfamily / 2-Layer Sandwich / Alpha BetaSimilarity search - Domain/homology SH3 domain / PH-like domain superfamily / 2-Layer Sandwich / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |   Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å MOLECULAR REPLACEMENT / Resolution: 1.5 Å |
|---|
 Authors Authors | Jaber Chehayeb, R. / Wang, J. / Stiegler, A.L. / Boggon, T.J. |
|---|
| Funding support |  United States, 2items United States, 2items | Organization | Grant number | Country |
|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | R01GM102262 |  United States United States | | American Heart Association | 19IPLOI34740007 |  United States United States |
|
|---|
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2020 Journal: J.Biol.Chem. / Year: 2020
Title: The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain.
Authors: Jaber Chehayeb, R. / Wang, J. / Stiegler, A.L. / Boggon, T.J. |
|---|
| History | | Deposition | Mar 26, 2020 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Jun 17, 2020 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 1, 2020 | Group: Database references / Category: citation / citation_author
Item: _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed ..._citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation_author.identifier_ORCID |
|---|
| Revision 1.2 | Aug 12, 2020 | Group: Database references / Category: citation
Item: _citation.journal_volume / _citation.page_first / _citation.page_last |
|---|
| Revision 1.3 | Oct 18, 2023 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords SIGNALING PROTEIN /
SIGNALING PROTEIN /  SH2 domain /
SH2 domain /  RasGAP /
RasGAP /  phosphopeptide /
phosphopeptide /  phosphotyrosine
phosphotyrosine Function and homology information
Function and homology information vasculogenesis / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases ...regulation of RNA metabolic process / regulation of actin filament polymerization / potassium channel inhibitor activity / negative regulation of cell adhesion / blood vessel morphogenesis / negative regulation of cell-matrix adhesion / mitotic cytokinesis / ephrin receptor signaling pathway /
vasculogenesis / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases ...regulation of RNA metabolic process / regulation of actin filament polymerization / potassium channel inhibitor activity / negative regulation of cell adhesion / blood vessel morphogenesis / negative regulation of cell-matrix adhesion / mitotic cytokinesis / ephrin receptor signaling pathway /  vasculogenesis / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / ruffle / EPHB-mediated forward signaling / phosphotyrosine residue binding / Downstream signal transduction /
vasculogenesis / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / ruffle / EPHB-mediated forward signaling / phosphotyrosine residue binding / Downstream signal transduction /  GTPase activator activity / VEGFR2 mediated cell proliferation / Regulation of RAS by GAPs /
GTPase activator activity / VEGFR2 mediated cell proliferation / Regulation of RAS by GAPs /  GTPase binding / regulation of cell shape / negative regulation of neuron apoptotic process / intracellular signal transduction /
GTPase binding / regulation of cell shape / negative regulation of neuron apoptotic process / intracellular signal transduction /  signaling receptor binding /
signaling receptor binding /  GTPase activity / negative regulation of apoptotic process /
GTPase activity / negative regulation of apoptotic process /  signal transduction /
signal transduction /  plasma membrane /
plasma membrane /  cytosol /
cytosol /  cytoplasm
cytoplasm
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å
MOLECULAR REPLACEMENT / Resolution: 1.5 Å  Authors
Authors United States, 2items
United States, 2items  Citation
Citation Journal: J.Biol.Chem. / Year: 2020
Journal: J.Biol.Chem. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6wax.cif.gz
6wax.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6wax.ent.gz
pdb6wax.ent.gz PDB format
PDB format 6wax.json.gz
6wax.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/wa/6wax
https://data.pdbj.org/pub/pdb/validation_reports/wa/6wax ftp://data.pdbj.org/pub/pdb/validation_reports/wa/6wax
ftp://data.pdbj.org/pub/pdb/validation_reports/wa/6wax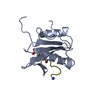

 10.15785/SBGRID/774 / Data set type: diffraction image data
10.15785/SBGRID/774 / Data set type: diffraction image data Links
Links Assembly
Assembly

 Movie
Movie Controller
Controller











 PDBj
PDBj









