| Entry | Database: PDB / ID: 6u1p
|
|---|
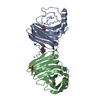
| Title | Crystal structure of VpsU (VC0916) from Vibrio cholerae |
|---|
 Components Components | Low molecular weight phosphotyrosine protein phosphatase |
|---|
 Keywords Keywords |  HYDROLASE / HYDROLASE /  Phosphatase / VC0916 / dimer Phosphatase / VC0916 / dimer |
|---|
| Function / homology | Protein-tyrosine phosphatase, low molecular weight /  Phosphotyrosine protein phosphatase I / Phosphotyrosine protein phosphatase I /  Phosphotyrosine protein phosphatase I superfamily / Low molecular weight phosphotyrosine protein phosphatase / Low molecular weight phosphatase family / Phosphotyrosine protein phosphatase I superfamily / Low molecular weight phosphotyrosine protein phosphatase / Low molecular weight phosphatase family /  protein-tyrosine-phosphatase / protein-tyrosine-phosphatase /  protein tyrosine phosphatase activity / protein tyrosine phosphatase activity /  Protein-tyrosine-phosphatase / Protein-tyrosine-phosphatase /  Protein-tyrosine-phosphatase Protein-tyrosine-phosphatase Function and homology information Function and homology information |
|---|
| Biological species |    Vibrio cholerae (bacteria) Vibrio cholerae (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.201 Å MOLECULAR REPLACEMENT / Resolution: 2.201 Å |
|---|
 Authors Authors | Tripathi, S.M. / Schwechheimer, C. / Herbert, K. / Osorio, J. / Yildiz, F.H. / Rubin, S.M. |
|---|
| Funding support |  United States, 1items United States, 1items | Organization | Grant number | Country |
|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | |  United States United States |
|
|---|
 Citation Citation |  Journal: Plos Pathog. / Year: 2020 Journal: Plos Pathog. / Year: 2020
Title: A tyrosine phosphoregulatory system controls exopolysaccharide biosynthesis and biofilm formation in Vibrio cholerae.
Authors: Schwechheimer, C. / Hebert, K. / Tripathi, S. / Singh, P.K. / Floyd, K.A. / Brown, E.R. / Porcella, M.E. / Osorio, J. / Kiblen, J.T.M. / Pagliai, F.A. / Drescher, K. / Rubin, S.M. / Yildiz, F.H. |
|---|
| History | | Deposition | Aug 16, 2019 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Aug 19, 2020 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Oct 7, 2020 | Group: Database references / Category: citation / citation_author
Item: _citation.country / _citation.journal_abbrev ..._citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year |
|---|
| Revision 1.2 | Oct 11, 2023 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ncs_dom_lim
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords HYDROLASE /
HYDROLASE /  Phosphatase / VC0916 / dimer
Phosphatase / VC0916 / dimer Phosphotyrosine protein phosphatase I /
Phosphotyrosine protein phosphatase I /  Phosphotyrosine protein phosphatase I superfamily / Low molecular weight phosphotyrosine protein phosphatase / Low molecular weight phosphatase family /
Phosphotyrosine protein phosphatase I superfamily / Low molecular weight phosphotyrosine protein phosphatase / Low molecular weight phosphatase family /  protein-tyrosine-phosphatase /
protein-tyrosine-phosphatase /  protein tyrosine phosphatase activity /
protein tyrosine phosphatase activity /  Protein-tyrosine-phosphatase /
Protein-tyrosine-phosphatase /  Protein-tyrosine-phosphatase
Protein-tyrosine-phosphatase Function and homology information
Function and homology information
 Vibrio cholerae (bacteria)
Vibrio cholerae (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.201 Å
MOLECULAR REPLACEMENT / Resolution: 2.201 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: Plos Pathog. / Year: 2020
Journal: Plos Pathog. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6u1p.cif.gz
6u1p.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6u1p.ent.gz
pdb6u1p.ent.gz PDB format
PDB format 6u1p.json.gz
6u1p.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/u1/6u1p
https://data.pdbj.org/pub/pdb/validation_reports/u1/6u1p ftp://data.pdbj.org/pub/pdb/validation_reports/u1/6u1p
ftp://data.pdbj.org/pub/pdb/validation_reports/u1/6u1p

 Links
Links Assembly
Assembly
 Movie
Movie Controller
Controller












 PDBj
PDBj