+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ogj | ||||||
|---|---|---|---|---|---|---|---|
| Title | MeCP2 MBD in complex with DNA | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA BINDING PROTEIN/DNA / MBD /  Structural Genomics / Structural Genomics /  Structural Genomics Consortium / SGC / DNA BINDING PROTEIN-DNA complex Structural Genomics Consortium / SGC / DNA BINDING PROTEIN-DNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationtrans-synaptic signaling by BDNF / regulation of action potential firing threshold / negative regulation of respiratory gaseous exchange / Loss of MECP2 binding ability to 5hmC-DNA / cellular response to isoquinoline alkaloid / positive regulation of anterograde dense core granule transport / positive regulation of retrograde dense core granule transport / positive regulation of branching morphogenesis of a nerve / catecholamine secretion / : ...trans-synaptic signaling by BDNF / regulation of action potential firing threshold / negative regulation of respiratory gaseous exchange / Loss of MECP2 binding ability to 5hmC-DNA / cellular response to isoquinoline alkaloid / positive regulation of anterograde dense core granule transport / positive regulation of retrograde dense core granule transport / positive regulation of branching morphogenesis of a nerve / catecholamine secretion / : / MECP2 regulates transcription of genes involved in GABA signaling / biogenic amine metabolic process / negative regulation of dendrite extension / principal sensory nucleus of trigeminal nerve development / cardiolipin metabolic process / negative regulation of locomotion involved in locomotory behavior / Loss of MECP2 binding ability to 5mC-DNA /  nervous system process involved in regulation of systemic arterial blood pressure / nervous system process involved in regulation of systemic arterial blood pressure /  proprioception / proprioception /  : / negative regulation of primary miRNA processing / negative regulation of smooth muscle cell differentiation / MECP2 regulates transcription of neuronal ligands / Loss of MECP2 binding ability to the NCoR/SMRT complex / negative regulation of dendritic spine development / Transcriptional Regulation by MECP2 / inositol metabolic process / regulation of respiratory gaseous exchange by nervous system process / double-stranded methylated DNA binding / thalamus development / : / negative regulation of primary miRNA processing / negative regulation of smooth muscle cell differentiation / MECP2 regulates transcription of neuronal ligands / Loss of MECP2 binding ability to the NCoR/SMRT complex / negative regulation of dendritic spine development / Transcriptional Regulation by MECP2 / inositol metabolic process / regulation of respiratory gaseous exchange by nervous system process / double-stranded methylated DNA binding / thalamus development /  genomic imprinting / positive regulation of microtubule nucleation / glucocorticoid metabolic process / genomic imprinting / positive regulation of microtubule nucleation / glucocorticoid metabolic process /  ventricular system development / cellular response to potassium ion / oligodendrocyte development / unmethylated CpG binding / phosphatidylcholine metabolic process / positive regulation of synaptic plasticity / respiratory gaseous exchange by respiratory system / olfactory bulb development / striatum development / positive regulation of dendrite extension / response to other organism / siRNA binding / neuron maturation / methyl-CpG binding / MECP2 regulates transcription factors / Loss of phosphorylation of MECP2 at T308 / response to ionizing radiation / lung alveolus development / spinal cord development / positive regulation of dendritic spine development / regulation of synapse organization / glutamine metabolic process / ventricular system development / cellular response to potassium ion / oligodendrocyte development / unmethylated CpG binding / phosphatidylcholine metabolic process / positive regulation of synaptic plasticity / respiratory gaseous exchange by respiratory system / olfactory bulb development / striatum development / positive regulation of dendrite extension / response to other organism / siRNA binding / neuron maturation / methyl-CpG binding / MECP2 regulates transcription factors / Loss of phosphorylation of MECP2 at T308 / response to ionizing radiation / lung alveolus development / spinal cord development / positive regulation of dendritic spine development / regulation of synapse organization / glutamine metabolic process /  startle response / dendrite development / startle response / dendrite development /  social behavior / negative regulation of blood vessel endothelial cell migration / Regulation of MECP2 expression and activity / negative regulation of astrocyte differentiation / behavioral fear response / glial cell proliferation / social behavior / negative regulation of blood vessel endothelial cell migration / Regulation of MECP2 expression and activity / negative regulation of astrocyte differentiation / behavioral fear response / glial cell proliferation /  heterochromatin / Nuclear events stimulated by ALK signaling in cancer / heterochromatin / Nuclear events stimulated by ALK signaling in cancer /  long-term memory / heterochromatin formation / MECP2 regulates neuronal receptors and channels / four-way junction DNA binding / positive regulation of glial cell proliferation / long-term memory / heterochromatin formation / MECP2 regulates neuronal receptors and channels / four-way junction DNA binding / positive regulation of glial cell proliferation /  Notch signaling pathway / sensory perception of pain / Notch signaling pathway / sensory perception of pain /  synapse assembly / synapse assembly /  excitatory postsynaptic potential / histone reader activity / cerebellum development / negative regulation of angiogenesis / molecular condensate scaffold activity / adult locomotory behavior / post-embryonic development / response to cocaine / long-term synaptic potentiation / promoter-specific chromatin binding / hippocampus development / response to lead ion / excitatory postsynaptic potential / histone reader activity / cerebellum development / negative regulation of angiogenesis / molecular condensate scaffold activity / adult locomotory behavior / post-embryonic development / response to cocaine / long-term synaptic potentiation / promoter-specific chromatin binding / hippocampus development / response to lead ion /  visual learning / visual learning /  protein localization / chromatin DNA binding / cerebral cortex development / protein localization / chromatin DNA binding / cerebral cortex development /  histone deacetylase binding / transcription corepressor activity / response to estradiol / histone deacetylase binding / transcription corepressor activity / response to estradiol /  gene expression / gene expression /  heart development / postsynapse / negative regulation of neuron apoptotic process / heart development / postsynapse / negative regulation of neuron apoptotic process /  nucleic acid binding / molecular adaptor activity / response to hypoxia / protein domain specific binding nucleic acid binding / molecular adaptor activity / response to hypoxia / protein domain specific bindingSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | ||||||
 Authors Authors | Lei, M. / Tempel, W. / Arrowsmith, C.H. / Bountra, C. / Edwards, A.M. / Min, J. / Structural Genomics Consortium (SGC) | ||||||
 Citation Citation |  Journal: Biochim Biophys Acta Gene Regul Mech / Year: 2019 Journal: Biochim Biophys Acta Gene Regul Mech / Year: 2019Title: Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain. Authors: Lei, M. / Tempel, W. / Chen, S. / Liu, K. / Min, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ogj.cif.gz 6ogj.cif.gz | 102.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ogj.ent.gz pdb6ogj.ent.gz | 73.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ogj.json.gz 6ogj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/og/6ogj https://data.pdbj.org/pub/pdb/validation_reports/og/6ogj ftp://data.pdbj.org/pub/pdb/validation_reports/og/6ogj ftp://data.pdbj.org/pub/pdb/validation_reports/og/6ogj | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6c1ySC  6ogkC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 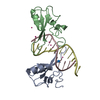
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  MECP2 / MeCp2 MECP2 / MeCp2Mass: 11179.503 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: MECP2 / Plasmid: pNIC-CH / Production host: Homo sapiens (human) / Gene: MECP2 / Plasmid: pNIC-CH / Production host:   Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3)-V2R-RIL / References: UniProt: P51608 Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3)-V2R-RIL / References: UniProt: P51608#2: DNA chain | | Mass: 3743.440 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) #3: DNA chain | | Mass: 3583.344 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) #4: Chemical | ChemComp-UNX / #5: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.84 Å3/Da / Density % sol: 33.01 % |
|---|---|
Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / Details: 30% PEG 2000 MME, 0.2M potassium bromide |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CLSI CLSI  / Beamline: 08ID-1 / Wavelength: 0.97949 Å / Beamline: 08ID-1 / Wavelength: 0.97949 Å | ||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: May 12, 2018 | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 0.97949 Å / Relative weight: 1 : 0.97949 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.8→45.09 Å / Num. obs: 19585 / % possible obs: 98.6 % / Redundancy: 3.3 % / CC1/2: 0.999 / Rmerge(I) obs: 0.047 / Rpim(I) all: 0.03 / Rrim(I) all: 0.056 / Net I/σ(I): 12 / Num. measured all: 65521 / Scaling rejects: 0 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: pdb entry 6c1y Resolution: 1.8→45.09 Å / Cor.coef. Fo:Fc: 0.956 / Cor.coef. Fo:Fc free: 0.946 / SU B: 7.441 / SU ML: 0.112 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.16 / ESU R Free: 0.144 Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : WITH TLS ADDED
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 78.24 Å2 / Biso mean: 37.548 Å2 / Biso min: 26.48 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.8→45.09 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.8→1.847 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj
















































