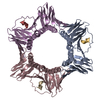
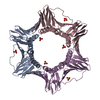
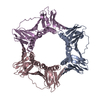
Entry Database : PDB / ID : 6k3aTitle Crystal structure of human PCNA in complex with DNMT1 PIP box motif. Peptide from DNA (cytosine-5)-methyltransferase 1 Proliferating cell nuclear antigen Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 2.3 Å Authors Jimenji, T. / Kori, S. / Arita, K. Funding support Organization Grant number Country Ministry of Education, Culture, Sports, Science and Technology (Japan) 18H02392 Japan Science and Technology 14530337
Journal : Biochem.Biophys.Res.Commun. / Year : 2019Title : Structure of PCNA in complex with DNMT1 PIP box reveals the basis for the molecular mechanism of the interaction.Authors : Jimenji, T. / Matsumura, R. / Kori, S. / Arita, K. History Deposition May 17, 2019 Deposition site / Processing site Revision 1.0 Jun 26, 2019 Provider / Type Revision 1.1 Jul 10, 2019 Group / Database references / Category Item _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year Revision 1.2 Jul 24, 2019 Group / Database references / Category Item / _citation.page_first / _citation.page_lastRevision 1.3 Nov 22, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accession
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords REPLICATION /
REPLICATION /  DNMT1 /
DNMT1 /  PCNA /
PCNA /  DNA methylation / PIP box
DNA methylation / PIP box Function and homology information
Function and homology information nuclear lamina / MutLalpha complex binding / Polymerase switching /
nuclear lamina / MutLalpha complex binding / Polymerase switching /  DNA (cytosine-5-)-methyltransferase / Telomere C-strand (Lagging Strand) Synthesis / Processive synthesis on the lagging strand /
DNA (cytosine-5-)-methyltransferase / Telomere C-strand (Lagging Strand) Synthesis / Processive synthesis on the lagging strand /  DNA (cytosine-5-)-methyltransferase activity /
DNA (cytosine-5-)-methyltransferase activity /  PCNA complex / DNA methylation-dependent heterochromatin formation / SUMOylation of DNA methylation proteins / Removal of the Flap Intermediate / Processive synthesis on the C-strand of the telomere / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / Transcription of E2F targets under negative control by DREAM complex / negative regulation of gene expression via chromosomal CpG island methylation / Polymerase switching on the C-strand of the telomere / Removal of the Flap Intermediate from the C-strand / female germ cell nucleus /
PCNA complex / DNA methylation-dependent heterochromatin formation / SUMOylation of DNA methylation proteins / Removal of the Flap Intermediate / Processive synthesis on the C-strand of the telomere / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / Transcription of E2F targets under negative control by DREAM complex / negative regulation of gene expression via chromosomal CpG island methylation / Polymerase switching on the C-strand of the telomere / Removal of the Flap Intermediate from the C-strand / female germ cell nucleus /  replisome / methyl-CpG binding / response to L-glutamate /
replisome / methyl-CpG binding / response to L-glutamate /  histone acetyltransferase binding / leading strand elongation / DNA polymerase processivity factor activity / G1/S-Specific Transcription / replication fork processing / response to dexamethasone / nuclear replication fork / SUMOylation of DNA replication proteins /
histone acetyltransferase binding / leading strand elongation / DNA polymerase processivity factor activity / G1/S-Specific Transcription / replication fork processing / response to dexamethasone / nuclear replication fork / SUMOylation of DNA replication proteins /  estrous cycle / PCNA-Dependent Long Patch Base Excision Repair / cyclin-dependent protein kinase holoenzyme complex /
estrous cycle / PCNA-Dependent Long Patch Base Excision Repair / cyclin-dependent protein kinase holoenzyme complex /  mismatch repair /
mismatch repair /  translesion synthesis / pericentric heterochromatin / response to cadmium ion /
translesion synthesis / pericentric heterochromatin / response to cadmium ion /  DNA polymerase binding /
DNA polymerase binding /  base-excision repair, gap-filling / positive regulation of vascular associated smooth muscle cell proliferation / epithelial cell differentiation / positive regulation of DNA repair / Translesion synthesis by REV1 / Translesion synthesis by POLK / Gap-filling DNA repair synthesis and ligation in GG-NER / Translesion synthesis by POLI /
base-excision repair, gap-filling / positive regulation of vascular associated smooth muscle cell proliferation / epithelial cell differentiation / positive regulation of DNA repair / Translesion synthesis by REV1 / Translesion synthesis by POLK / Gap-filling DNA repair synthesis and ligation in GG-NER / Translesion synthesis by POLI /  DNA methylation / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest /
DNA methylation / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest /  replication fork / positive regulation of DNA replication / PRC2 methylates histones and DNA / male germ cell nucleus / Defective pyroptosis /
replication fork / positive regulation of DNA replication / PRC2 methylates histones and DNA / male germ cell nucleus / Defective pyroptosis /  liver regeneration / promoter-specific chromatin binding / nuclear estrogen receptor binding / Recognition of DNA damage by PCNA-containing replication complex / cellular response to amino acid stimulus / Termination of translesion DNA synthesis / Translesion Synthesis by POLH / NoRC negatively regulates rRNA expression / HDR through Homologous Recombination (HRR) / Dual Incision in GG-NER /
liver regeneration / promoter-specific chromatin binding / nuclear estrogen receptor binding / Recognition of DNA damage by PCNA-containing replication complex / cellular response to amino acid stimulus / Termination of translesion DNA synthesis / Translesion Synthesis by POLH / NoRC negatively regulates rRNA expression / HDR through Homologous Recombination (HRR) / Dual Incision in GG-NER /  receptor tyrosine kinase binding / cellular response to hydrogen peroxide / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / cellular response to UV / cellular response to xenobiotic stimulus / response to estradiol / E3 ubiquitin ligases ubiquitinate target proteins /
receptor tyrosine kinase binding / cellular response to hydrogen peroxide / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / cellular response to UV / cellular response to xenobiotic stimulus / response to estradiol / E3 ubiquitin ligases ubiquitinate target proteins /  heart development /
heart development /  chromosome, telomeric region / damaged DNA binding /
chromosome, telomeric region / damaged DNA binding /  nuclear body / negative regulation of gene expression /
nuclear body / negative regulation of gene expression /  centrosome / DNA-templated transcription /
centrosome / DNA-templated transcription /  chromatin binding /
chromatin binding /  chromatin / protein-containing complex binding / positive regulation of gene expression / negative regulation of transcription by RNA polymerase II /
chromatin / protein-containing complex binding / positive regulation of gene expression / negative regulation of transcription by RNA polymerase II /  enzyme binding /
enzyme binding /  DNA binding /
DNA binding /  RNA binding / extracellular exosome / zinc ion binding
RNA binding / extracellular exosome / zinc ion binding
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å
MOLECULAR REPLACEMENT / Resolution: 2.3 Å  Authors
Authors Japan, 2items
Japan, 2items  Citation
Citation Journal: Biochem.Biophys.Res.Commun. / Year: 2019
Journal: Biochem.Biophys.Res.Commun. / Year: 2019 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6k3a.cif.gz
6k3a.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6k3a.ent.gz
pdb6k3a.ent.gz PDB format
PDB format 6k3a.json.gz
6k3a.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/k3/6k3a
https://data.pdbj.org/pub/pdb/validation_reports/k3/6k3a ftp://data.pdbj.org/pub/pdb/validation_reports/k3/6k3a
ftp://data.pdbj.org/pub/pdb/validation_reports/k3/6k3a
 Links
Links Assembly
Assembly
 Components
Components / PCNA / Cyclin
/ PCNA / Cyclin
 Homo sapiens (human) / Gene: PCNA / Production host:
Homo sapiens (human) / Gene: PCNA / Production host: 
 Escherichia coli (E. coli) / References: UniProt: P12004
Escherichia coli (E. coli) / References: UniProt: P12004

 Homo sapiens (human) / References: UniProt: K7ERQ1, UniProt: P26358*PLUS
Homo sapiens (human) / References: UniProt: K7ERQ1, UniProt: P26358*PLUS Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  Photon Factory
Photon Factory  / Beamline: BL-17A / Wavelength: 0.98 Å
/ Beamline: BL-17A / Wavelength: 0.98 Å : 0.98 Å / Relative weight: 1
: 0.98 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller









 PDBj
PDBj


















